Modifying or Creating an Association File¶
Learning Goals¶
This Notebook is designed to walk the user (you) through: Creating or altering the association (asn) file used by the Cosmic Origins Spectrograph (COS) pipeline to determine which data to process:
1. Examining an association file
2. Editing an existing association file
- 2.1. Removing an exposure
+ 2.1.1. Removing a bad exposure
+ 2.1.2. Filtering to a single exposure (e.g. for LP6 data)
- 2.2. Adding an exposure
3. Creating an entirely new association file
- 3.1. Simplest method
- 3.2. With fits header metadata
- 3.3. Association files for non-TAGFLASH datasets
+ 3.3.1. Gathering the exposure information
0. Introduction¶
The Cosmic Origins Spectrograph (COS) is an ultraviolet spectrograph on-board the Hubble Space Telescope (HST) with capabilities in the near ultraviolet (NUV) and far ultraviolet (FUV).
This tutorial aims to prepare you to alter the association file used by the CalCOS pipeline. Association files are fits files containing a binary table extension, which list their "member" files: science and calibration exposures which the pipeline will process together into spectral data products.
- For an in-depth manual to working with COS data and a discussion of caveats and user tips, see the COS Data Handbook.
- For a detailed overview of the COS instrument, see the COS Instrument Handbook.
We'll demonstrate creating an asn file in three ways: First, we'll demonstrate editing an existing asn file to add or remove an exposure. Second, we'll show how to create an entirely new asn file. Finally, we'll show an example of creating an asn file for use with SPLIT wavecal data, such as that obtained at COS lifetime position 6 (LP6).
We will import the following packages:¶
numpyto handle array functionsastropy.io fitsandastropy.table Tablefor accessing FITS filesglob,os, andshutilfor working with system filesglobhelps us search for filenamesosandshutilfor moving files and deleting folders, respectively
astroquery.mast MastandObservationsfor finding and downloading data from the MAST archivedatetimefor updating fits headers with today's datepathlib Pathfor managing system paths
If you have an existing astroconda environment, it may or may not already have the necessary packages to run this Notebook. To create a Python environment capable of running all the data analyses in these COS Notebooks, please see Section 1 of our Notebook tutorial on setting up an environment.
# Import for: array manipulation
import numpy as np
# Import for: data table manipulatioon
import pandas as pd
# Import for: reading fits files
from astropy.io import fits
from astropy.table import Table
# Import for: system files
import glob
import os
import shutil
# Import for: downloading the data
from astroquery.mast import Observations
# Import for: changing modification date in a fits header
import datetime
#Import for: working with system paths
from pathlib import Path
We will also define a few directories we will need:¶
# These will be important directories for the Notebook
datadir = Path('./data/')
outputdir = Path('./output/')
plotsdir = Path('./output/plots/')
# Make the directories if they don't already exist
datadir.mkdir(exist_ok=True), outputdir.mkdir(exist_ok=True), plotsdir.mkdir(exist_ok=True)
print("Made the following directories:"+"\n ", f'./{datadir}, ./{outputdir}, ./{plotsdir}')
And we will need to download the data we wish to filter and analyze¶
We choose the exposures with the association obs_ids: ldif01010 and ldif02010 because we happen to know that some of the exposures in these groups failed, which gives us a real-world use case for editing an association file. Both ldif01010 and ldif02010 are far-ultraviolet (FUV) datasets on the quasi-stellar object (QSO) PDS 456.
For more information on downloading COS data, see our Notebook tutorial on downloading COS data.
pl = Observations.get_product_list(Observations.query_criteria(obs_id='ldif0*10')) # search for the correct obs_ids and get the product list
fpl = Observations.filter_products(pl,
productSubGroupDescription=['RAWTAG_A', 'RAWTAG_B','ASN']) # filter to rawtag and asn files in the product list
Observations.download_products(fpl, download_dir=str(datadir)) # Download these chosen products
for gfile in glob.glob("**/ldif*/*.fits", recursive=True): # Move all fits files in this set to the base data directory
os.rename(gfile,datadir / os.path.basename(gfile))
shutil.rmtree(datadir / 'mastDownload') # Delete the now-empty, nested mastDownload directory
1. Examining an association file¶
Association files are lists of "member" files - science and wavelength calibration exposures - which the CalCOS pipeline will process together.
Above, we downloaded two association files and the rawtag data files which are their members. We will begin by searching for the association files and reading one of them (with observation ID LDIF01010). We could just as easily pick ldif02010.
asnfiles = sorted(glob.glob("**/*ldif*asn*", recursive=True)) # There will be two (ldif01010_asn.fits and ldif02010_asn.fits)
asnfile = asnfiles[0] # We want to work primarily with ldif01010_asn.fits
asn_contents = Table.read(asnfile) # Gets the contents of the asn file
asn_contents # Display these contents
The association file has three columns: MEMNAME, MEMTYPE, MEMPRSNT which we describe below. More information can be found in Section 3 of the COS Data Handbook.
MEMNAME(short for "member name"): The rootname of the file, e.g.ldif01u0qfor the fileldif01u0q_rawtag_a.fitsMEMTYPE("membership type"): Whether the item is a science exposure (EXP-FP), a wavecal exposure (EXP-SWAVE/EXP-GWAVE/EXP-AWAVE), or the output product (PROD-FP)MEMPRSNT("member present"): Whether to include the file inCalCOS' processing
We also see that this association file has five rows: four science exposures denoted with the MEMTYPE = EXP-FP, and an output product with MEMTYPE = PROD-FP.
In the cell below, we examine a bit about each of the exposures as a diagnostic:
for memname, memtype in zip(asn_contents['MEMNAME'], asn_contents["MEMTYPE"]): # Cycles through each file in asn table
memname = memname.lower() # Find file names in lower case letters
if memtype == 'EXP-FP': # We only want to look at the exposure files
rt_a = (glob.glob(f"**/*{memname}*rawtag_a*", recursive=True))[0] # Find the actual filepath of the memname for rawtag_a and rawtag_b
rt_b = (glob.glob(f"**/*{memname}*rawtag_b*", recursive=True))[0]
# Now print all these diagnostics:
print(f"Association {(fits.getheader(rt_a))['ASN_ID']} has {memtype} exposure {memname.upper()} with \
exposure time {(fits.getheader(rt_a, ext=1))['EXPTIME']} seconds at cenwave {(fits.getheader(rt_a, ext=0))['CENWAVE']} \
Å and FP-POS {(fits.getheader(rt_a, ext=0))['FPPOS']}.")
We notice that something seems amiss with the science exposure LDIF01TYQ: This file has an exposure time of 0.0 seconds - something has gone wrong. In this case, there was a guide star acquisition failure as described on the data preview page.
In the next section, we will correct this lack of data by removing the bad exposure and combining in exposures from the other association group.
2. Editing an existing association file¶
2.1. Removing an exposure¶
2.1.1. Removing a bad exposure¶
We know that at least one of our exposures - ldif01tyq - is not suited for combination into the final product. It has an exposure time of 0.0 seconds, in this case from a guide star acquisition failure. This is a generalizable issue, as you may often know an exposure is "bad" for many reasons: perhaps it was taken with the shutter closed, or with anomolously high background noise, or any number of reasons we may wish to exclude an exposure from our data. To do this, we will need to alter our existing association file before we re-run CalCOS. Afterwards we will compare the flux of the resulting spectrum made without the bad exposure to the same dataset with the bad exposure. The flux levels should match closely.
We again see the contents of our main association file below. Note that True/False and 1/0 are essentially interchangable in the MEMPRSNT column.
Table.read(asnfiles[0])
We can set the MEMPRSNT value to False or 0 for our bad exposure. If we were to run the CalCOS pipeline on the edited association file, it would not be processed. CalCOS can take a long time to run. To avoid slowing down this Notebook, the code to run CalCOS on the association files we have created has been spun out into Python files in this directory with the names test_<type of association file>.py (in this case, test_removed_exposure_asn.py). If you wish to re-run the analysis within, you will need to specify a valid cache of CRDS reference files (see our Setup Notebook for details on downloading reference files) in the Python file.
with fits.open(asnfile, mode='update') as hdulist: # We need to change values with the asnfile opened and in 'update' mode
tbdata = hdulist[1].data # This is where the table data is read into
for expfile in tbdata: # Check if each file is one of the bad ones
if expfile['MEMNAME'] in ['LDIF01TYQ']:
expfile['MEMPRSNT'] = False # If so, set MEMPRSNT to False AKA 0
# Copy this file, which we will go on to edit, in case we want to run CalCOS on it.
shutil.copy(asnfile, datadir / "removed_badfile_asn.fits")
# Re-read the table to see the change
Table.read(asnfile)
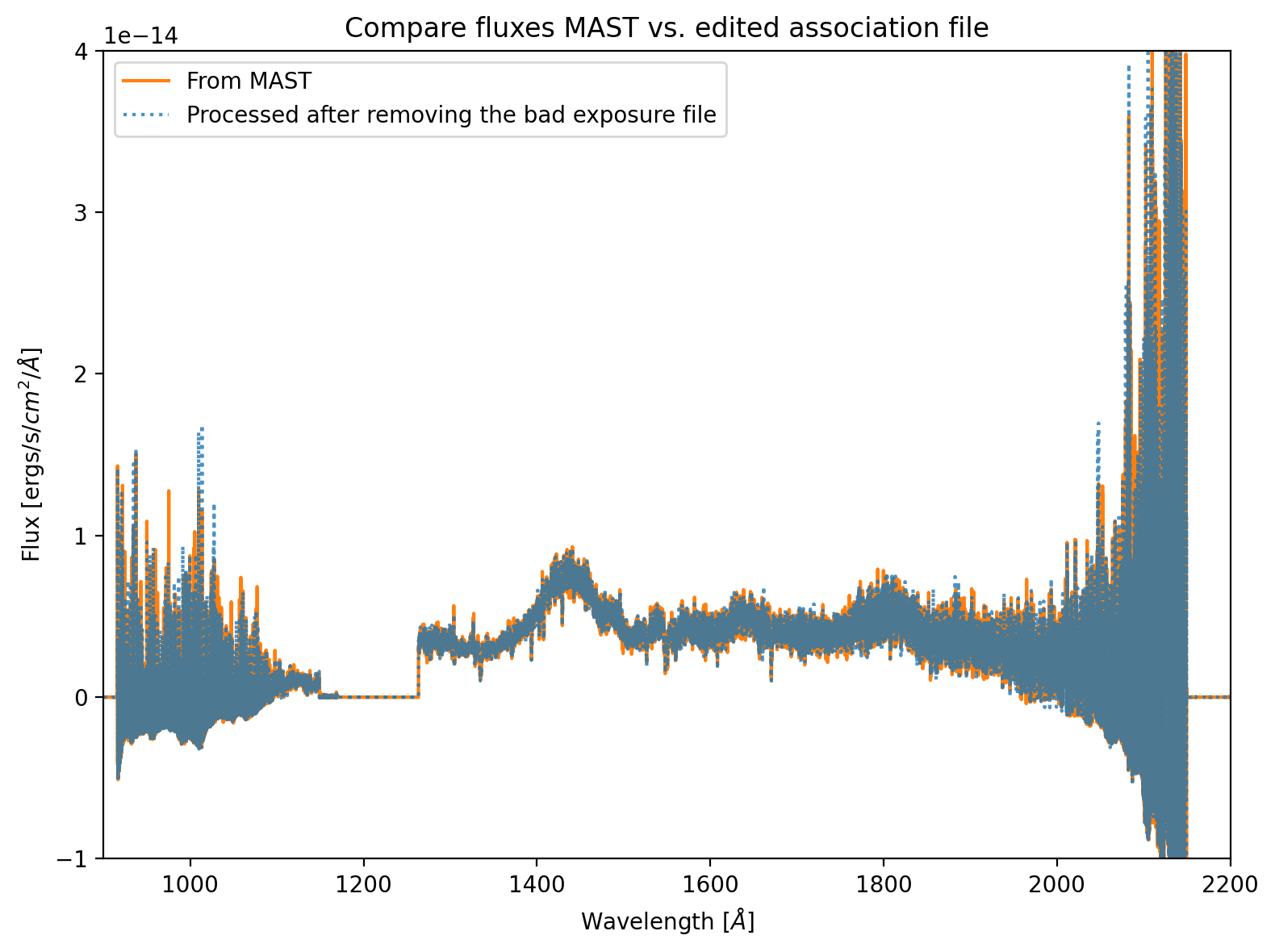
To show what would happen if we run CalCOS on this association file, we include Figure 1. This figure shows that the flux level of the spectrum retrieved from MAST matches that which was processed after removing the bad exposure. Slight differences in the two spectra may arise from MAST using a different random seed value. What matters for this comparison is that the fluxes match well, as they should because the empty exposure adds 0 counts over 0 time. Thus, for the purposes of flux calibration, it should be ignored. Other types of bad exposures, for instance those taken with the source at the edge of COS's aperture, would impact the fluxes.
Figure 1. Comparison of fluxes between the data retrieved from MAST, and the same data reprocessed after removing the bad exposure¶
2.1.2. Filtering to a single exposure (e.g. for LP6 data)¶
Another situation when users often wish to remove exposures from an association file is when re-processing individual exposures taken at COS's lifetime position 6 (LP6) or other modes at which the wavelength calibration data is not contained by the science exposures' rawtag files.
Often users wish to process a single exposure's file with CalCOS. With the TAGFLASH wavelength calibration method standard at all lifetime positions prior to LP6, the wavelength calibration data was taken concurrently with the science data and was contained by the same rawtag data files. However, as described in this COS 2030 plan poster, this is not possible at LP6. Instead, separate wavelength calibration exposures are taken at separate lifetime positions in a process known as SPLIT wavecals). As a result, the wavelength calibration data no longer exists in the same file as the science data for such exposures.
For TAGFLASH data, users could run CalCOS directly on their rawtag file of interest (though using an association file was always recommended). However, doing so with LP6 data would not allow the proper wavelength calibration. Instead, users should create a custom association file created by removing exposures from the default association file. We demonstrate this below.
We'll alter an LP6 association file with the observation ID letc01010.
Let's begin by downloading the default LP6 association file from MAST and displaying it:
# Search for the correct obs_ids and get the list of data products
lp6_observation_products = Observations.get_product_list(
Observations.query_criteria(obs_id='letc01010'),
)
# Download the asnfile product list, then get the 0th element of the resulting path list, since there's only 1 asn file
lp6_original_asnfile = Path(
Observations.download_products(
Observations.filter_products(
lp6_observation_products,
productSubGroupDescription=['ASN'],
),
download_dir=str(datadir)
)['Local Path'][0]
)
# Show the default association file for the LP6 dataset.
Table.read(lp6_original_asnfile)
As expected, we see 4 EXP-FP members, which are the science exposures at COS's 4 fixed-pattern noise positions (FP-POS). Each is bracketed by a SPLIT wavecal wavelength calibration exposure on each side (EXP-SWAVE).
If we only wish to process one of the science exposures, (along with its wavelength calibration exposures,) we turn off all the other exposures by setting their MEMPRSNT values to 0/False. The product file (PROD-FP) must also be turned on, and should be renamed to show that it will only include data from a single exposure. Below, we do so, assuming we wish to process only the FP3 science exposure: LETC01MTQ (and its wavelength calibration exposures: LETC01M6Q and LETC01MVQ).
exposures_to_use = ['LETC01M6Q','LETC01MTQ','LETC01MVQ'] # WAVECAL -> SCIENCE -> WAVECAL
print(f"We will turn off all the input exposures except for: {exposures_to_use}." +\
" We will also leave the output product file turned on.")
# Make a copy of the original asn file with a name to indicate it will only process a single chosen exposure
lp6_1exposure_asnfile = shutil.copy(lp6_original_asnfile, datadir / 'letc01mtq_only_asn.fits')
# Turn off all the other exposures in the copy
with fits.open(lp6_1exposure_asnfile, mode='update') as hdulist:
tbdata = hdulist[1].data
for expfile in tbdata:
# Turn off files except those listed here
if expfile['MEMNAME'] not in exposures_to_use:
expfile['MEMPRSNT'] = False # If so, set MEMPRSNT to False AKA 0
# Turn on and rename the product file to indicate it will only include the chosen exposure
if expfile['MEMTYPE'] == 'PROD-FP':
expfile['MEMPRSNT'] = True # Turn on the product file
expfile['MEMNAME'] = "LETC01MTQ_only" # Rename the product file
# Re-read the table to see the change
Table.read(lp6_1exposure_asnfile)
2.2. Adding an exposure¶
In section 2.1.1, we removed the failed exposure taken with FP-POS = 1 from our ldif01010 association file. When possible, we usually want to combine one or more of each of the four FP-POS types. In this example scenario, let's add the FP-POS = 1 exposure from the other association group. Please note that combining data from separate visits with different target acquisitions can result in true errors which are higher than those calculated by CalCOS.
We combine the datasets here as an example only and do not specifically endorse combining these exposures.
Also note that you should only combine files taken using the same grating and central wavelength settings in this manner.
In the cell below, we determine which exposure from LDIF02010 was taken with FP-POS = 1.
- It does this by looping through the files listed in
LDIF02010's association file, and then reading in that file's header to check if itsFPPOSvalue equals 1. - It also prints some diagnostic information about all of the exposure files.
asn_contents_2 = Table.read(asnfiles[1]) # Reads the contents of the 2nd asn file
for memname, memtype in zip(asn_contents_2['MEMNAME'], asn_contents_2["MEMTYPE"]): # Loops through each file in asn table for `LDIF02010`
memname = memname.lower() # Convert file names to lower case letters, as in actual filenames
if memtype == 'EXP-FP': # We only want to look at the exposure files
rt_a = (glob.glob(f"**/*{memname}*rawtag_a*", recursive=True))[0] # Search for the actual filepath of the memname for rawtag_a
rt_b = (glob.glob(f"**/*{memname}*rawtag_b*", recursive=True))[0] # Search for the actual filepath of the memname for rawtag_b
# Now print all these diagnostics:
print(f"Association {(fits.getheader(rt_a))['ASN_ID']} has {memtype} exposure {memname.upper()} with \
exptime {(fits.getheader(rt_a, ext=1))['EXPTIME']} seconds at cenwave {(fits.getheader(rt_a, ext=0))['CENWAVE']} Å and FP-POS {(fits.getheader(rt_a, ext=0))['FPPOS']}.")
if (fits.getheader(rt_a, ext=0))['FPPOS'] == 1:
print(f"^^^ The one above this has the FP-POS we are looking for ({memname.upper()})^^^\n")
asn2_fppos1_name = memname.upper() # Save the right file basename in a variable
There's a slightly different procedure to add a new exposure to the list rather than remove one.
Here we will read the table in the fits association file into an astropy Table. We can then add a row into the right spot, filling it with the new file's MEMNAME, MEMTYPE, and MEMPRSNT. Finally, we have to save this table into the existing fits association file.
asn_orig_table = Table.read(asnfile) # Read in original data from the file
asn_orig_table.insert_row(len(asn_orig_table)- 1 , [asn2_fppos1_name,'EXP-FP',1]) # Add a row with the right name after all the original EXP-FP's
new_table = fits.BinTableHDU(asn_orig_table) # Turn this into a fits Binary Table HDU
with fits.open(asnfile, mode='update') as hdulist: # We need to change data with the asnfile opened and in 'update' mode
hdulist[1].data = new_table.data # Change the orig file's data to the new table data we made
print(f"Added {asn2_fppos1_name} to the association file.")
Now, we can see there is a new row with our exposure from the other asn file group: LDIF02NMQ.
Table.read(asnfile)
Excellent. In the next section we will create a new association file from scratch.
3. Creating an entirely new association file¶
Users will rarely need to create an entirely new association file. Much more often, they should alter an existing one found on MAST. However, a user may wish to create a new association file if they are combining altered exposures, perhaps after using the costools.splittag tool. This section describes how to make one from scratch.
For the sake of demonstration, we will generate a new association file with four exposure members: even-numbered FP-POS (2,4) from the first original association (LDIF01010), and odd-numbered FP-POS (1,3) from from the second original association (LDIF02010).
From section 2, we see that this corresponds to :
| Name | Original asn | FP-POS |
|---|---|---|
| LDIF02010 | LDIF02NMQ | 1 |
| LDIF01010 | LDIF01U0Q | 2 |
| LDIF02010 | LDIF02NUQ | 3 |
| LDIF01010 | LDIF01U4Q | 4 |
3.1. Simplest method¶
Below, we manually build up an association file from the three necessary columns (MEMNAME, MEMTYPE, MEMPRSNT).
# Adding the exposure file details to the association table
new_asn_memnames = ['LDIF02NMQ','LDIF01U0Q','LDIF02NUQ','LDIF01U4Q'] # MEMNAME
types = ['EXP-FP', 'EXP-FP', 'EXP-FP', 'EXP-FP'] # MEMTYPE
included = [True, True, True, True] # MEMPRSNT
# Adding the ASN details to the end of the association table
new_asn_memnames.append('ldifcombo'.upper()) # MEMNAME column
types.append('PROD-FP') # MEMTYPE column
included.append(True) # MEMPRSNT column
# Putting together the fits table
# 40 is the number of characters allowed in this field with the MEMNAME format = 40A.
# If your rootname is longer than 40, you will need to increase this
c1 = fits.Column(name='MEMNAME', array=np.array(new_asn_memnames), format='40A')
c2 = fits.Column(name='MEMTYPE', array=np.array(types), format='14A')
c3 = fits.Column(name='MEMPRSNT', format='L', array=included)
asn_table = fits.BinTableHDU.from_columns([c1, c2, c3])
# Writing the fits table
asn_table.writeto(outputdir / 'ldifcombo_asn.fits', overwrite=True)
print('Saved '+ 'ldifcombo_asn.fits'+ f" in the output directory: {outputdir}")
Examining the file we have created:
We see that the data looks correct - exactly the table we want!
Table.read(outputdir / 'ldifcombo_asn.fits')
However, the 0th and 1st fits headers no longer contain useful information about the data.
While CalCOS will process these files, vital header keys may not be propagated. If it's important to preserve the header information, you may follow the steps in the next section.
fits.getheader(outputdir / 'ldifcombo_asn.fits', ext=0)
fits.getheader(outputdir / 'ldifcombo_asn.fits', ext=1)
3.2. With fits header metadata¶
We can instead build up a new file with our old file's fits header, and alter it to reflect our changes.
We first build a new association file, a piecewise combination of our original file's headers and our new table:
with fits.open(asnfile, mode='readonly') as hdulist: # Open up the old asn file
hdulist.info() # Shows the first hdu is empty except for the header we want
hdu0 = hdulist[0] # We want to directly copy over the old 0th header/data-unit AKA "hdu":
# essentially a section of data and its associated metadata, called a "header"
# see https://fits.gsfc.nasa.gov/fits_primer.html for info on fits structures
d0 = hdulist[0].data # gather the data from the header/data unit to allow the readout
h1 = hdulist[1].header # gather the header from the 1st header/data unit to copy to our new file
hdu1 = fits.BinTableHDU.from_columns([c1, c2, c3], header=h1) # Put together new 1st hdu from old header and new data
new_HDUlist = fits.HDUList([hdu0,hdu1]) # New HDUList from old HDU 0 and new combined HDU 1
new_HDUlist.writeto(outputdir / 'ldifcombo_2_asn.fits', overwrite=True) # Write this out to a new file
new_asnfile = outputdir / 'ldifcombo_2_asn.fits' # Path to this new file
print('\nSaved '+ 'ldifcombo_2_asn.fits'+ f"in the output directory: {outputdir}")
Now we edit the relevant values in our fits headers that are different from the original.
Note: It is possible that a generic fits file may have different values you may wish to change. It is highly recommended to examine your fits headers.
date = datetime.date.today() # Find today's date
# Below, make a dict of what header values we want to change, corresponding to [new value, extension the value lives in, 2nd extension if applies]
keys_to_change = {'DATE':[f'{date.year}-{date.month}-{date.day}',0], 'FILENAME':['ldifcombo_2_asn.fits',0],
'ROOTNAME':['ldifcombo_2',0,1], 'ASN_ID':['ldifcombo_2',0], 'ASN_TAB':['ldifcombo_2_asn.fits',0], 'ASN_PROD':['False',0],
'EXTVER':[2,1], 'EXPNAME':['ldifcombo_2',1]}
# Actually change the values below (verbosely):
for keyval in keys_to_change.items():
print(f"Editing {keyval[0]} in Extension {keyval[1][1]}")
fits.setval(filename=new_asnfile, keyword=keyval[0], value=keyval[1][0], ext=keyval[1][1])
# Below is necessary as some keys are repeated in both headers ('ROOTNAME')
if len(keyval[1])>2:
print(f"Editing {keyval[0]} in Extension {keyval[1][2]}")
fits.setval(filename=new_asnfile, keyword=keyval[0], value= keyval[1][0], ext=keyval[1][2])
And now we have created our new association file. The file is now ready to be used in the CalCOS pipeline!
If you're interested in testing your file by running it through the CalCOS pipeline, you may wish to run the test_asn.py file included in this subdirectory of the GitHub repository. i.e. from the command line:
$ python test_asn.py
Note that you must first...
- Have created the file by running this Notebook
- Alter line 21 of
test_asn.pyto set the lref directory to wherever you have your cache of CRDS reference files (see our Setup Notebook).
If the test runs successfully, it will create a plot in the subdirectory ./output/plots/ .
3.3. Association files for non-TAGFLASH datasets¶
As touched upon in Section 2.1.2., most COS exposures contain the calibration lamp data necessary to perform wavelength calibration; however, exposures taken at LP6 and those taken with AUTO and GO wavecals) (including BOA and ACCUM mode exposures) have separate exposures for wavelength calibration. Table 1 below summarizes the types of wavelength calibration modes. More information may be found in Section 5.7 of the COS Instrument Handbook.
Table 1. Comparison of wavelength calibration modes¶
| Wavelength calibration mode | TAGFLASH |
SPLIT Wavecal |
AUTO Wavecal |
GO Wavecal | |
|---|---|---|---|---|---|
| When used | Default for LP1-LP5 | Default for LP6 | Used for LP1-LP5 non-TAGFLASH exposures | User specified (rare) | |
| Description | Wavelength calibration lamp flashes concurrently with science exposure | Wavelength calibration lamp flashes at another LP | Wavelength calibration lamp flashes separately from science exposure | Wavelength calibration lamp flashes separately from science exposure | |
| Additional wavelength calibration file in the association table | None - wavelength calibration data included in the EXP-FPs of the science exposures |
EXP-SWAVE |
EXP-AWAVE |
EXP-GWAVE |
Non-TAGFLASH datasets must be processed from a single association file which includes all the science and wavelength calibration exposures, properly labeled with the correct MEMTYPEs.
Users will rarely need to build an entirely new association file for non-TAGFLASH datasets. The most common reason for a user to have non-TAGFLASH data is that the exposures were obtained using the SPLIT wavecal method at LP6. The association files for these files are correctly created and made available in the MAST archive. Removing specific exposures can most easily be done as in Section 2.1.2. However, if a user needed to craft a completely customized CalCOS run, for instance to combine multiple non-TAGFLASH datasets, they can follow this section.
In this section we will manually recreate the association file for the SPLIT wavecal data from the same dataset whose association file we altered as part of Section 2.1.2. We could just as easily do so with AUTO or GO wavecal data. We will first select and examine the relevant exposures, then we will combine them into an association file.
3.3.1. Gathering the exposure information¶
We begin by downloading the data from a visit which utilized COS' LP6 split wavecal mode (proposal ID: 16907, observation ID: letc01010). We previously downloaded the association file for this dataset in Section 2.1.2, but here we download the rest of the data products we will need.
# Search for the correct obs_ids and get the list of data products
lp6_observation_products = Observations.get_product_list(
Observations.query_criteria(obs_id='letc01010'),
)
# Filter and download the rawtag files and asn file in the product list from MAST
# First filter to just the rawtag products
lp6_rawtags_pl = Observations.filter_products(
lp6_observation_products,
productSubGroupDescription=['RAWTAG_A', 'RAWTAG_B'],
)
# Download these chosen rawtag products
lp6_rawtags = Observations.download_products(lp6_rawtags_pl, download_dir=str(datadir))['Local Path']
We'll begin by taking a look at the original association file from MAST in the cell below. Note that the exposures are ordered in groups of a single EXP-FP (also known as science or EXTERNAL/SCI) exposure bracketed on both sides by an EXP-SWAVE (SPLIT wavecal) exposure (i.e. wavecal-->science-->wavecal). This is often the case for non-TAGFLASH files, especially for exposures longer than ~600 seconds.
Table.read(lp6_original_asnfile)
We'll simulate an occasion on which we would need to create our own association file by supposing we want to process only data from this observation taken in the FP1 or FP3 configuration. We could achieve the same result by taking the default association file from MAST and turning the MEMPRSNT value to False for all the FP2 and FP4 files, similarly to in Section 2.1.2.
Below, we filter to such exposures and check whether they meet our expectations for SPLIT wavecal data. Namely, we check that:
- The files are listed in chronological order.
- All science (
EXTERNAL/SCI) exposures are bracketed by aWAVECALexposure on either side. - The data is organized into groups of 3 exposures with the grouping described in test 2 (
WAVECAL-->EXTERNAL/SCI-->WAVECAL).
CalCOS does not strictly need the files to be arranged in chronological order; however, it is a helpful exercise to make sure we include all the files we need to calibrate this dataset. We can investigate the files manually and diagnose any failures in the table printed by the following cell.
# Filter the rawtag files to exposures at FP1 and FP3
lp6_rawtags_fp13 = [rt for rt in lp6_rawtags if fits.getval(rt, "FPPOS", ext=0) in [1,3]]
# Gather information on all the exposures
rawtag_a_exptypes_dict = {rt : fits.getval(rt, "EXPTYPE", ext=0) for rt in lp6_rawtags_fp13 if "rawtag_a" in rt}
rawtag_a_rootnames = [fits.getval(rt, "ROOTNAME", ext=0) for rt in lp6_rawtags_fp13 if "rawtag_a" in rt]
rawtag_a_exptypes = [fits.getval(rt, "EXPTYPE", ext=0) for rt in lp6_rawtags_fp13 if "rawtag_a" in rt]
rawtag_a_expstart_times = [fits.getval(rt, "EXPSTART", ext=1) for rt in lp6_rawtags_fp13 if "rawtag_a" in rt]
# Test 1
# Check chronological sorting
assert all(np.diff(rawtag_a_expstart_times) >= 0),"These exposures are not ordered by start time."
print("Passed Test #1: The exposures are in chronological order.")
# Test 2
# Check science exposures are bracketed by wavecals
for i, rta_type in enumerate(rawtag_a_exptypes):
if rta_type == "EXTERNAL/SCI":
assert rawtag_a_exptypes[i-1] == "WAVECAL", "EXTERNAL/SCI exposure not preceded by a WAVECAL exposure"
assert rawtag_a_exptypes[i+1] == "WAVECAL", "EXTERNAL/SCI exposure not followed by a WAVECAL exposure"
print("Passed Test #2: Each EXTERNAL/SCI is bracketted on both sides by a WAVECAL exposure.")
# Test 3
# Check that only these groups of exposures exist (WAVECAL-->EXTERNAL/SCI-->WAVECAL)
for group_of_3 in [rawtag_a_exptypes[3*i:3*i+3] for i in range(int(len(rawtag_a_exptypes)/3))]:
assert group_of_3 == ['WAVECAL', 'EXTERNAL/SCI', 'WAVECAL'], "Incorrect groupings of exposures"
print("Passed Test #3: No unexpected groupings of files were found.")
Below, we may inspect these files by eye, and see that they are indeed ordered correctly.
pd.DataFrame({
"Rootname":[name.upper() for name in rawtag_a_rootnames],
"Exposure_type":rawtag_a_exptypes,
"Exposure_start_date":rawtag_a_expstart_times, # Date in MJD
"Seconds_since_first_exposure":\
# convert time since the first exposure into seconds
86400*np.subtract(rawtag_a_expstart_times, min(rawtag_a_expstart_times))
})
Once we are sure that the correct exposures are selected, we gather the information we need for an association table:
- The exposure ID (
ROOTNAME) of each exposure.- When capitalized, this is the same as the
MEMNAMEwe find in association files.
- When capitalized, this is the same as the
- The exposure type (
EXPTYPE) of each exposure.- We will need to convert from the way that exposure types are written in the FITS header to the way
CalCOSwill recognize them in the association file.
- We will need to convert from the way that exposure types are written in the FITS header to the way
To prevent duplication, we only gather this information from either the rawtag_a or rawtag_b files.
# Choose either rawtag_a (default) of rawtag_b files if no rawtag_a files found
if any(["rawtag_a" in rt for rt in lp6_rawtags_fp13]):
segment_found="a"
elif any(["rawtag_b" in rt for rt in lp6_rawtags_fp13]):
segment_found="b"
else:
print("Neither rawtag_a nor rawtag_b found.")
lp6_fp13_memnames = [fits.getval(rt, "ROOTNAME", ext=0).upper() for rt in lp6_rawtags_fp13 if f"rawtag_{segment_found}" in rt]
lp6_fp13_exptypes = [fits.getval(rt, "EXPTYPE", ext=0) for rt in lp6_rawtags_fp13 if f"rawtag_{segment_found}" in rt]
# We need to change the wavecals' MEMTYPE to "EXP-SWAVE", and the sciences' to "EXP-FP"
# If these were GO or AUTO wavecals, we would instead convert to "EXP-AWAVE" or "EXP-GWAVE"
lp6_fp13_exptypes = [
"EXP-SWAVE" if etype == "WAVECAL"
else "EXP-FP" if etype == "EXTERNAL/SCI"
else None for etype in lp6_fp13_exptypes
]
print(f"Gathered exposure information for creating a new non-TAGFLASH association file.")
3.3.2. Creating the SPLIT wavecal association file¶
Now that we've gathered MEMNAMEs and MEMTYPEs of our exposures, we can create the association file. This very closely mirrors Section 3.2.
# Adding the exposure file details to the association table
new_asn_memnames = lp6_fp13_memnames # MEMNAME
types = lp6_fp13_exptypes # MEMTYPE
included = [True] * len(lp6_fp13_memnames) # MEMPRSNT
# Adding the output science product details to the end of the association table columns
new_asn_memnames.append('splitwave'.upper()) # MEMNAME column
types.append('PROD-FP') # MEMTYPE column
included.append(True) # MEMPRSNT column
# Convert the columns into a the necessary form for a fits file
c1 = fits.Column(name='MEMNAME', array=np.array(new_asn_memnames), format='40A')
c2 = fits.Column(name='MEMTYPE', array=np.array(types), format='14A')
c3 = fits.Column(name='MEMPRSNT', format='L', array=included)
with fits.open(lp6_original_asnfile, mode='readonly') as hdulist: # Open up the old asn file
hdulist.info() # Shows the first hdu is empty except for the header we want
hdu0 = hdulist[0] # We want to directly copy over the old 0th header/data-unit
d0 = hdulist[0].data # gather the data from the header/data unit to allow the readout
h1 = hdulist[1].header # gather the header from the 1st header/data unit to copy to our new file
hdu1 = fits.BinTableHDU.from_columns([c1, c2, c3], header=h1) # Put together new 1st hdu from old header and new data
new_HDUlist = fits.HDUList([hdu0,hdu1]) # New HDUList from old HDU 0 and new combined HDU 1
new_HDUlist.writeto(outputdir / 'splitwave_asn.fits', overwrite=True) # Write this out to a new file
new_asnfile = outputdir / 'splitwave_asn.fits' # Path to this new file
print('\nSaved '+ 'splitwave_asn.fits'+ f" in the output directory: {outputdir}")
Table.read(outputdir/"splitwave_asn.fits")
Well done - you have now successfully created the association file you need to process your SPLIT wavecal data. If you wish to test it by running the CalCOS data calibration pipeline on it, you may run the file test_splitwave_asn.py in this directory. Note the following requirements:
test_splitwave_asn.pycan only be run after creating theasnfiles in Section 3.3 of this Notebook.- Your version of
CalCOSmust be ≥ 3.4. This version introduced the ability to process SPLIT wavecal data to the pipeline. You can check your version withcalcos --versionfrom the command line. - You must first update the
lrefenvironment variable set intest_splitwave_asn.pyto the path to a directory containing all of your reference files. For more information on setting up such a directory, see our Notebook on setting up an environment for COS data analysis.
About this Notebook¶
Author: Nat Kerman: nkerman@stsci.edu
Contributors: Elaine Mae Frazer, Travis Fischer
Updated On: 2022-09-20
This tutorial was generated to be in compliance with the STScI style guides and would like to cite the Jupyter guide in particular.
Citations¶
If you use astropy, matplotlib, astroquery, or numpy for published research, please cite the
authors. Follow these links for more information about citations:
