BOTS Time Series Observations#
Use case: Bright Object Time Series; extracting exoplanet spectra.
Data: JWST simulated NIRSpec data from ground-based campaign; GJ436b spectra from the Goyal et al. (2018).
Tools: scikit, lmfit, scipy, matplotlip, astropy, pandas.
Cross-intrument: .
Documentation: This notebook is part of a STScI’s larger post-pipeline Data Analysis Tools Ecosystem.
Author: David K. Sing (dsing@jhu.edu)
Last updated: 2 July 2020
Introduction#
This notebook uses time series JWST NIRSpec data taken during a ground-based campaign to illustrate extracting exoplanet spectra from time-series observations.
The data are derived from the ISIM-CV3, the cryovacuum campaign of the JWST Integrated Science Instrument Module (ISIM), that took place at Goddard Space Flight Center during the winter 2015-2016 (Kimble et al. 2016). The data can be found at https://www.cosmos.esa.int/web/jwst-nirspec/test-data, and detailed and insightful report of the data by G. Giardino, S. Birkmann, P. Ferruit, B. Dorner, B. Rauscher can be found here: ftp://ftp.cosmos.esa.int/jwstlib/ReleasedCV3dataTimeSeries/CV3_TimeSeries_PRM.tgz
This NIRSpec time series dataset has had a transit light curve injected at the pixel-level, which closely mimics a bright object time series (BOTS) observation of a transiting exoplanet. In this case, a GJ436b spectra from the Goyal et al. (2018) exoplanet grid was selected (clear atmosphere at solar metallicity). With an actual NIRSpec dataset, the noise properties of the detector, jitter, and the effects on extracting exoplanet spectra from time-series observations can more accurately simulated.
Broadly the aim of this notebook is to work with these time series observations to:
Extract 1D spectra from the 2D spectral images.
Define a time series model to fit to the wavelength dependent transit light curve.
Fit each time series wavelength bin of the 1D spectra, measuring the desired quantity \(R_{pl}(\lambda)/R_{star}\).
Produce a measured transmission spectrum that can then be compared to models.
The example outputs the fit light curves for each spectral bin, along with fitting statistics.
Load packages#
This notebook uses packages (matplotlib, astropy, scipy, glob, lmfit, pickle, os, sklearn) which can all be installed in a standard fashion through pip.
Several routines to calculate limb-darkening and a transit model were extracted from ExoTiC-ISm (Laginja & Wakeford 2020 ; https://github.com/hrwakeford/ExoTiC-ISM), and slightly adapted. The full set of stellar models used for the limb-darkening calculation can also be downloaded from ExoTiC-ISM, as this notebook only downloads and loads the single stellar model used to generate the limb darkening.
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from matplotlib.backends.backend_pdf import PdfPages
from astropy.utils.data import download_file
from astropy.table import Table
from astropy.io import fits, ascii
from astropy.modeling.models import custom_model
from astropy.modeling.fitting import LevMarLSQFitter
from scipy.interpolate import interp1d, splev, splrep
from scipy.io import readsav
from scipy import stats
import glob
import lmfit
import pickle
from os import path, mkdir
from sklearn.linear_model import LinearRegression
import pandas as pd
import os
import shutil
Setup Parameters#
Parameters of the fit include directories where the data and limb darkening stellar models are held, along with properties of the planet and star. The stellar and planet values that have been entered here (modeled after GJ436) are the same as was used to model the injected transit. Note, the 4500K stellar model used to inject the transit was hotter than GJ436A.
# SETUP ----------------------------------------------
# Setup directories
save_directory = './notebookrun2/' # Local directory to save files to
data_directory = './' # Local data to work with fits files if desired
# Setup Detector Properties & Rednoise measurement timescale
gain = 1.0 # 2D spectra has already converted to counts, gain of detector is 1.0
binmeasure = 256 # Binning technique to measure rednoise, choose bin size to evaluate sigma_r
number_of_images = 8192 # Number of images in the dataset
# Setup Planet Properties
grating = 'NIRSpecPrism'
ld_model = '3D' # 3D/1D stellar model choice (transit was injected with the 3D model)
# Setup Stellar Properties for Limb-Darkening Calculation
Teff = 4500 # Effective Temperature (K)
logg = 4.5 # Surface Gravity
M_H = 0.0 # Stellar Metallicity log_10[M/H]
Rstar = 0.455 # Planet radius (in units of solar radii Run)
# Setup Transit parameters (can get from NASA exoplanet archive)
t0 = 2454865.084034 # bjd time of inferior conjunction
per = 2.64389803 # orbital period (days) BJD_TDB
rp = 0.0804 # Planet radius (in units of stellar radii)
a_Rs = 14.54 # Semi-major axis (input a/Rstar so units of stellar radii)
inc = 86.858 * (2*np.pi/360) # Orbital inclination (in degrees->radians)
ecc = 0.0 # Eccentricity
omega = 0.0 * (2*np.pi/360) # Longitude of periastron (in degrees->radians)
rho_star = (3*np.pi)/(6.67259E-8*(per*86400)**2)*(a_Rs)**3 # Stellar Density (g/cm^3) from a/Rs
# a_Rs=(rho_star*6.67259E-8*per_sec*per_sec/(3*np.pi))**(1/3) # a/Rs from Stellar Density (g/cm^3)
# Create local directories
if not path.exists(save_directory):
mkdir(save_directory) # Create a new directory to save outputs to if needed
if not path.exists(save_directory+'3DGrid'):
mkdir(save_directory+'3DGrid') # Create new directory to save
limb_dark_directory = save_directory # Point to limb darkeing directory contaning 3DGrid/ directory
Download and Load NIRSpec data#
The fits images are loaded, and information including the image date and science spectra are saved.
A default flux offset value BZERO is also taken from the header and subtracted from every science frame.
Reading in the 2^13 fits files is slow. To speed things up, we created a pickle file of the for first instance the fits images are loaded. This 1GB pickle file is loaded instead of reading the fits files if found.
Alternatively, the fits files can be downloaded here: https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/Archive.Trace_SLIT_A_1600_SRAD-PRM-PS-6007102143_37803_JLAB88_injected.tar.gz. The images are in a tar.gz archvie, which needs to be un-archived and data_directory variable set to the directory in the SETUP cell above.
The cell below downloads the 1GB JWST data pickle file, and several other files needed.
# Download 1GB NIRSpec Data
fn_jw = save_directory+'jwst_data.pickle'
if not path.exists(fn_jw):
fn = download_file('https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/jwst_data.pickle')
dest = shutil.move(fn, save_directory+'jwst_data.pickle')
print('JWST Data Download Complete')
# Download further files needed, move to local directory for easier repeated access
fn_sens = download_file('https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/NIRSpec.prism.sensitivity.sav')
dest = shutil.move(fn_sens, save_directory+'NIRSpec.prism.sensitivity.sav') # Move files to save_directory
fn_ld = download_file('https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/3DGrid/mmu_t45g45m00v05.flx')
destld = shutil.move(fn_ld, save_directory+'3DGrid/mmu_t45g45m00v05.flx')
JWST Data Download Complete
Loads the Pickle File data. Alternatly, the data can be read from the original fits files.
if path.exists(fn_jw):
dbfile = open(fn_jw, 'rb') # for reading also binary mode is important
jwst_data = pickle.load(dbfile)
print('Loading JWST data from Pickle File')
bjd = jwst_data['bjd']
wsdata_all = jwst_data['wsdata_all']
shx = jwst_data['shx']
shy = jwst_data['shy']
common_mode = jwst_data['common_mode']
all_spec = jwst_data['all_spec']
exposure_length = jwst_data['exposure_length']
dbfile.close()
print('Done')
elif not path.exists(fn_jw):
# Load all fits images
# Arrays created for BJD time, and the white light curve total_counts
list = glob.glob(data_directory+"*.fits")
index_of_images = np.arange(number_of_images)
bjd = np.zeros((number_of_images))
exposure_length = np.zeros((number_of_images))
all_spec = np.zeros((32, 512, number_of_images))
for i in index_of_images:
img = list[i]
print(img)
hdul = fits.open(img)
# hdul.info()
bjd_image = hdul[0].header['BJD_TDB']
BZERO = hdul[0].header['BZERO'] # flux value offset
bjd[i] = bjd_image
expleng = hdul[0].header['INTTIME'] # Total integration time for one MULTIACCUM (seconds)
exposure_length[i] = expleng/86400. # Total integration time for one MULTIACCUM (days)
print(bjd[i])
data = hdul[0].data
# total counts in image
# total_counts[i]=gain*np.sum(data[11:18,170:200]-BZERO) # total counts in 12 pix wide aperture around pixel 60 image
all_spec[:, :, i] = gain * (data-BZERO) # Load all spectra into an array subtract flux value offset
hdul.close()
# Sort data
srt = np.argsort(bjd) # index to sort
bjd = bjd[srt]
# total_counts=total_counts[srt]
exposure_length = exposure_length[srt]
all_spec[:, :, :] = all_spec[:, :, srt]
# Get Wavelength of Data
file_wave = download_file('https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/JWST_NIRSpec_wavelength_microns.txt')
f = open(file_wave, 'r')
wsdata_all = np.genfromtxt(f)
print('wsdata size :', wsdata_all.shape)
print('Data wavelength Loaded :', wsdata_all)
print('wsdata new size :', wsdata_all.shape)
# Read in Detrending parameters
# Mean of parameter must be 0.0 to be properly normalized
# Idealy standard deviation of parameter = 1.0
file_xy = download_file('https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/JWST_NIRSpec_Xposs_Yposs_CM_detrending.txt')
f = open(file_xy, 'r')
data = np.genfromtxt(f, delimiter=',')
shx = data[:, 0]
shy = data[:, 1]
common_mode = data[:, 2]
# Store Data in a pickle file
jwst_data = {'bjd': bjd, 'wsdata_all': wsdata_all, 'shx': shx, 'shy': shy, 'common_mode': common_mode, 'all_spec': all_spec, 'exposure_length': exposure_length}
dbfile = open('jwst_data.pickle', 'ab') # Its important to use binary mode
pickle.dump(jwst_data, dbfile)
dbfile.close()
Loading JWST data from Pickle File
Done
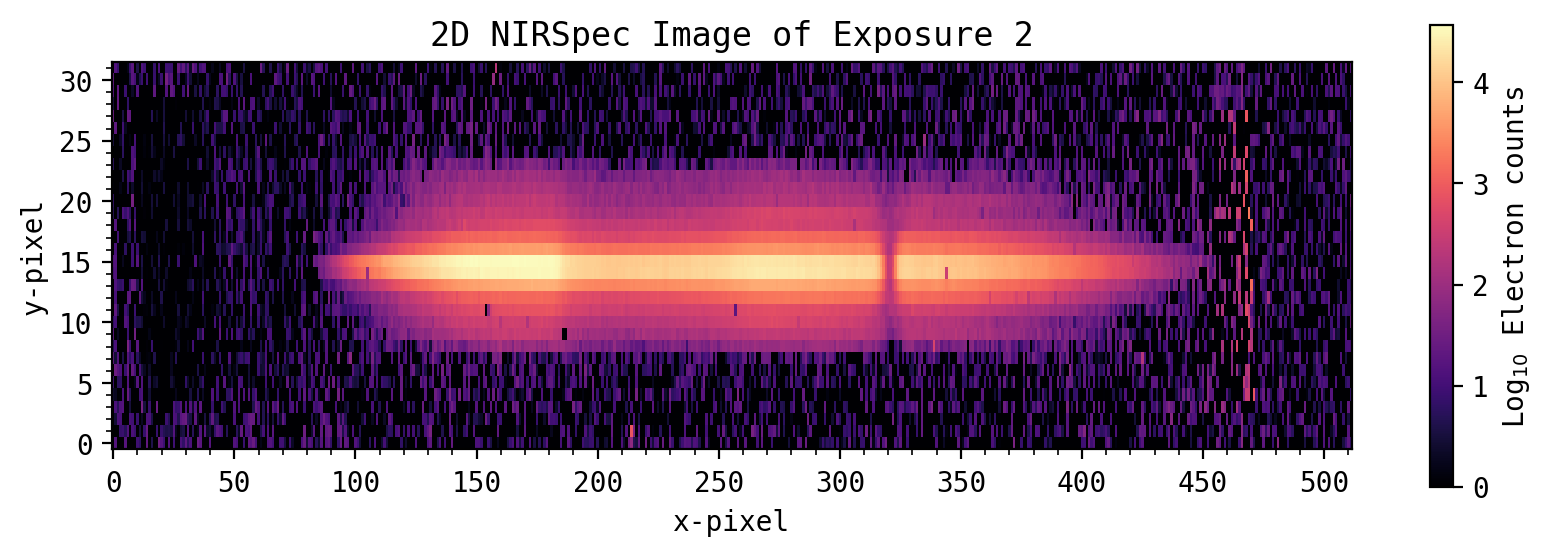
Visualizing the 2D spectral data#
expnum = 2 # Choose Exposure number to view
plt.rcParams['figure.figsize'] = [10.0, 3.0] # Figure dimensions
plt.rcParams['figure.dpi'] = 200 # Resolution
plt.rcParams['savefig.dpi'] = 200
plt.rcParams['image.aspect'] = 5 # Aspect ratio (the CCD is quite long!!!)
plt.cmap = plt.cm.magma
plt.cmap.set_bad('k', 1.)
plt.rcParams['image.cmap'] = 'magma' # Colormap
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['font.family'] = "monospace"
plt.rcParams['font.monospace'] = 'DejaVu Sans Mono'
img = all_spec[:, :, expnum]
zeros = np.where(img <= 0) # Plot on a log scale, so set zero or negative values to a small number
img[zeros] = 1E-10
fig, axs = plt.subplots()
f = axs.imshow(np.log10(img), vmin=0) # Plot image
plt.xlabel('x-pixel')
plt.ylabel('y-pixel')
axs.yaxis.set_major_locator(ticker.MultipleLocator(5))
axs.yaxis.set_minor_locator(ticker.MultipleLocator(1))
axs.xaxis.set_major_locator(ticker.MultipleLocator(50))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(10))
plt.title('2D NIRSpec Image of Exposure ' + str(expnum))
fig.colorbar(f, label='Log$_{10}$ Electron counts', ax=axs)
plt.show()
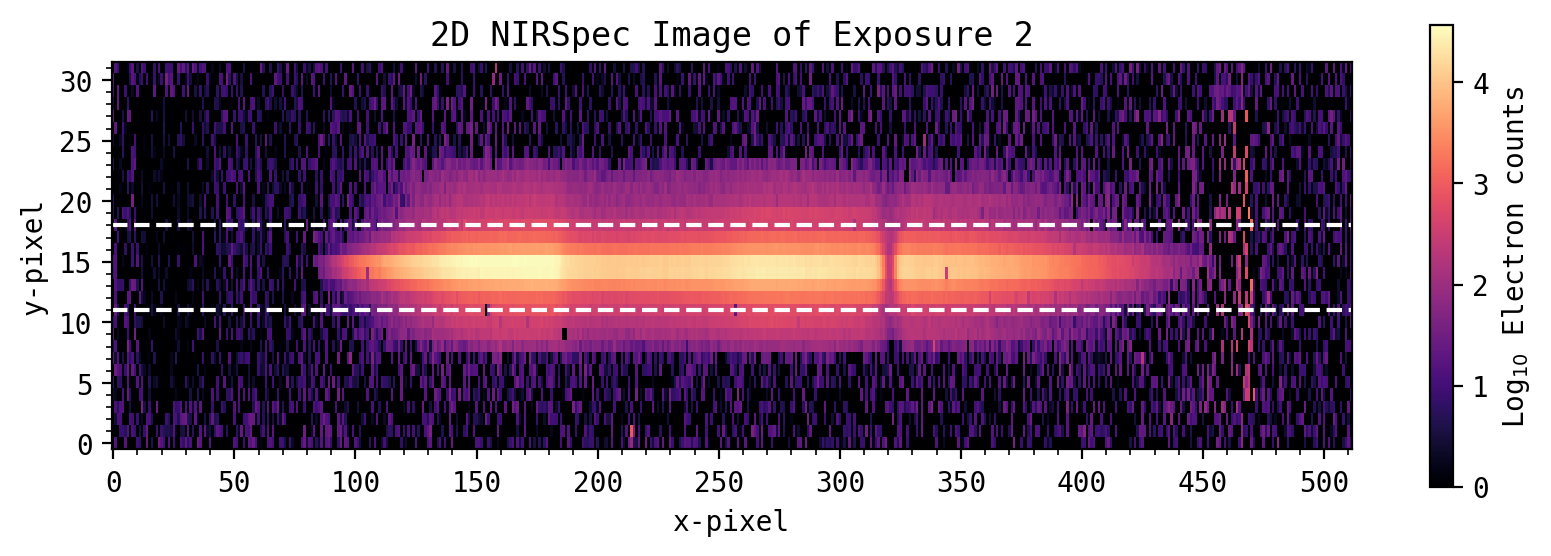
Extract 1D spectra from 2D array of images#
Ideally, extracting 1D spectra from the 2D images would use optimal aperture extraction along a fit trace with routines equivalent to IRAF/apall. This functionality is not yet available in astro-py.
Several processing steps have already been applied. The 2D spectra here have been flat field corrected, and 1/f noise has been removed from each pixel by subtracting the median count rate from the un-illuminated pixels along each column (see Giardino et al. for more information about 1/f noise). Each 2D image has also been aligned in the X and Y directions, such that each pixel corresponds to the same wavelength. As the CV3 test had no requirements for flux stability, the ~1% flux variations from the LED have also been removed.
For spectral extraction, the example here simply uses a simple summed box. The 8192 2D spectra have been pre-loaded into a numpy array. The spectra peaks at pixel Y=16. For each column, an aperature sum is taken over Y-axis pixels 11 to 18, which contains most of the spectrum counts. Wider aperture would add more counts, but also introduces more noise.
Further cleaning steps are not done here
Ideally, the pixels flagged as bad for various reasons should be cleaned.
Cosmic rays should be identified and removed.
all_spec.shape
y_lower = 11 # Lower extraction aperture
y_upper = 18 # Upper extraction aperture
all_spec_1D = np.sum(all_spec[y_lower:y_upper, :, :], axis=0) # Sum along Y-axis from pixels 11 to 18
# Plot
plt.rcParams['figure.figsize'] = [10.0, 3.0] # Figure dimensions
plt.rcParams['figure.dpi'] = 200 # Resolution
plt.rcParams['savefig.dpi'] = 200
plt.rcParams['image.aspect'] = 5 # Aspect ratio (the CCD is quite long!!!)
plt.cmap = plt.cm.magma
plt.cmap.set_bad('k', 1.)
plt.rcParams['image.cmap'] = 'magma' # Colormap
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['font.family'] = "monospace"
plt.rcParams['font.monospace'] = 'DejaVu Sans Mono'
img = all_spec[:, :, expnum]
zeros = np.where(img <= 0) # Plot on a log scale, so set zero or negative values to a small number
img[zeros] = 1E-10
fig, axs = plt.subplots()
f = axs.imshow(np.log10(img), vmin=0) # Plot image
plt.xlabel('x-pixel')
plt.ylabel('y-pixel')
axs.yaxis.set_major_locator(ticker.MultipleLocator(5))
axs.yaxis.set_minor_locator(ticker.MultipleLocator(1))
axs.xaxis.set_major_locator(ticker.MultipleLocator(50))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(10))
plt.axhline(y_lower, color='w', ls='dashed')
plt.axhline(y_upper, color='w', ls='dashed')
plt.title('2D NIRSpec Image of Exposure '+str(expnum))
fig.colorbar(f, label='Log$_{10}$ Electron counts', ax=axs)
plt.show()
Visualizing the 1D spectral data#
fig, axs = plt.subplots()
f = plt.plot(wsdata_all, all_spec_1D[:, 0], linewidth=2, zorder=0) # overplot Transit model at data
plt.xlabel(r'Wavelength ($\mu$m)')
plt.ylabel('Flux (e-)')
axs.xaxis.set_major_locator(ticker.MultipleLocator(0.5))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(0.1))
plt.annotate('H$_2$O', xy=(3.0, 42000))
plt.annotate('CO$_2$', xy=(4.2, 42000))
plt.show()
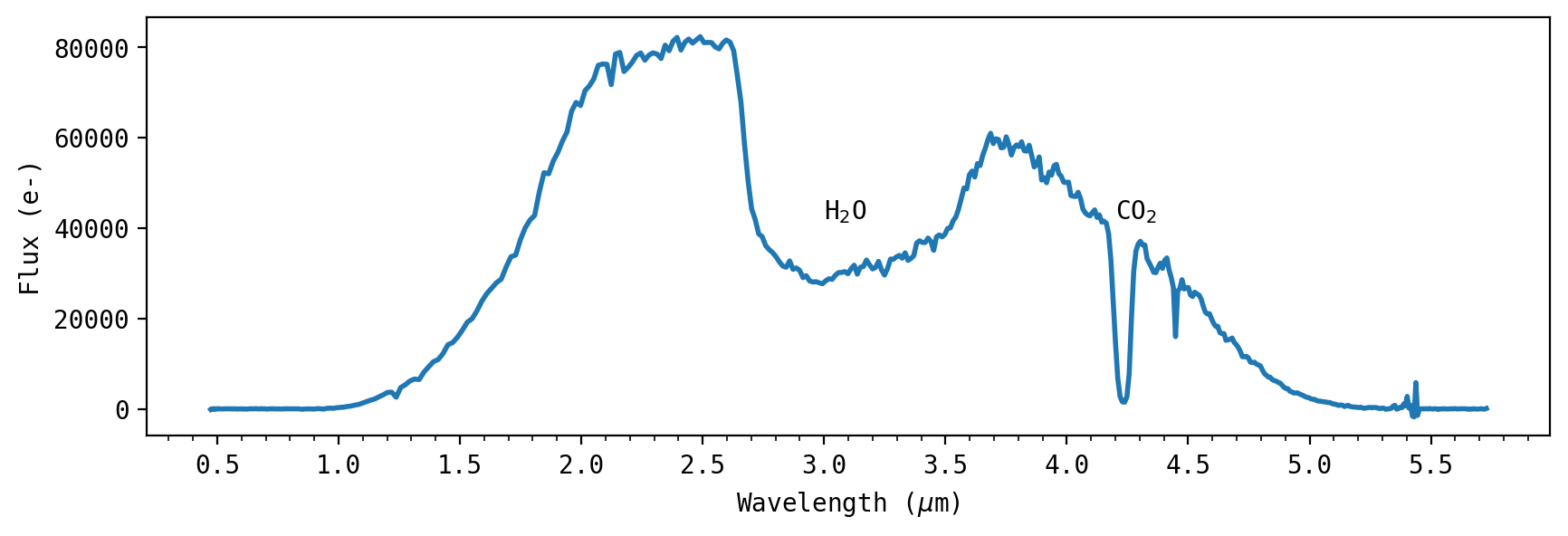
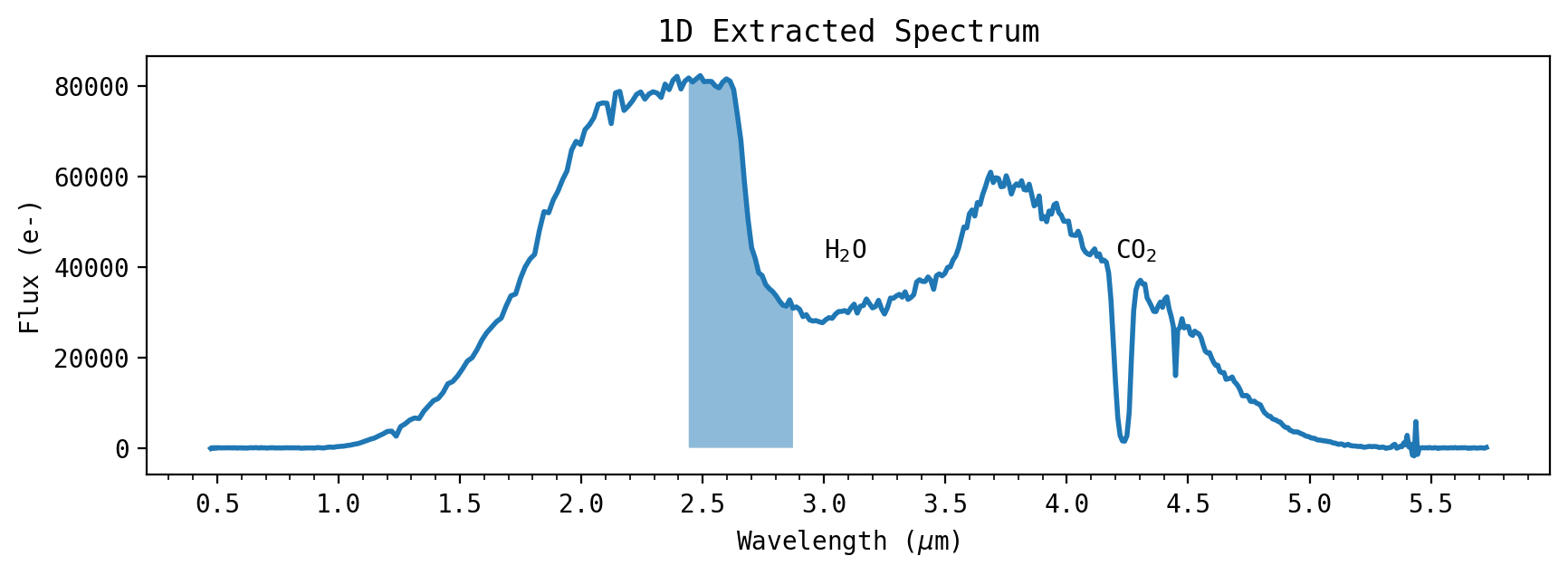
The CV3 test observed a lamp with a similar PSF as JWST will have, and has significant counts from about 1.5 to 4.5 \(\mu\)m.
The cryogenic test chamber had CO\(_2\) and H\(_2\)O ice buildup on the window, which can be seen as spectral absorption features in the 2D spectra.
Calculate Orbital Phase and a separate fine grid model used for plotting purposes#
# Calculate Orbital Phase
phase = (bjd-t0) / (per) # phase in days relative to T0 ephemeris
phase = phase - np.fix(phase[number_of_images-1]) # Have current phase occur at value 0.0
t_fine = np.linspace(np.min(bjd), np.max(bjd), 1000) # times at which to calculate light curve
phase_fine = (t_fine-t0)/(per) # phase in days relative to T0 ephemeris
phase_fine = phase_fine-np.fix(phase[number_of_images-1]) # Have current phase occur at value 0.0
b0 = a_Rs * np.sqrt((np.sin(phase * 2 * np.pi)) ** 2 + (np.cos(inc) * np.cos(phase * 2 * np.pi)) ** 2)
intransit = (b0-rp < 1.0E0).nonzero() # Select indicies between first and fourth contact
outtransit = (b0-rp > 1.0E0).nonzero() # Select indicies out of transit
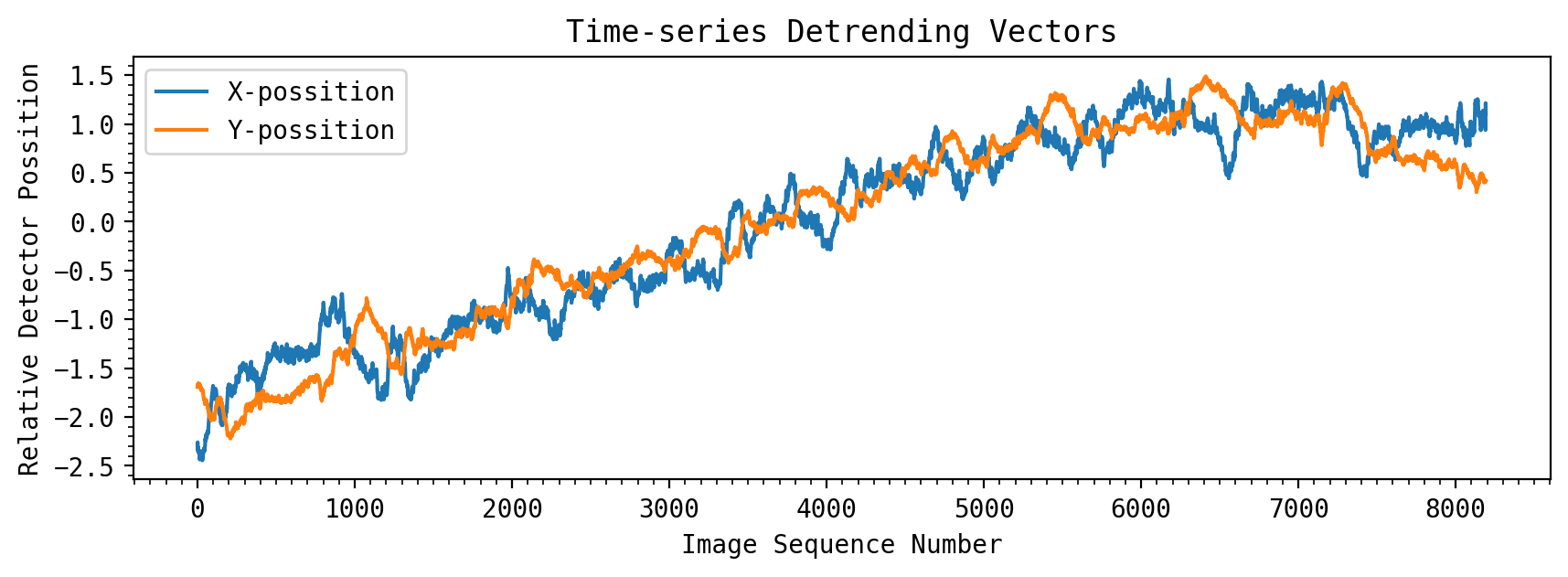
Dealing with Systematic Drift On the Detector#
The CV3 test assessed the stability of the instrument by introducing a large spatial jitter and drift. This resulted in a significant X,Y movement of the spectra on the 2D detector. While this bulk shift has been removed which aligns the spectra, intra- and inter- pixel sensitivities introduce flux variations which need to be removed. The jitter from the CV3 test was more than 30 mas, which is ~4X larger than the JWST stability requirement. Thus, in orbit these detector effects are expected to be significantly smaller, but they will still be present and will need to be modeled and removed from time series observations.
The detector X, Y positions here were measured from cross-correlation of the 2D images (collapsing the spectra along one dimension first), and are saved in arrays \(shx\) and \(shy\). These detending vectors would ideally be measured using the trace position values from the spectral extraction of each integration, as that could also accurately measure integration-to-integration how the spectra spatially changed on the detector.
The detector shifts have original amplitudes near 0.2 pixels, though the vectors have had initial normalization. For detrending purposes, these arrays should have a mean of 0 and standard deviation of 1.0.
A residual color-dependent trend with the LED lamp can also been seen in the CV3 data, which can be partly removed by scaling original common-mode lamp trend, which was measured using the CV3 white light curve.
shx_tmp = shx / np.mean(shx) - 1.0E0 # Set Mean around 0.0
shx_detrend = shx_tmp/np.std(shx_tmp) # Set standard deviation to 1.0
shy_tmp = shy / np.mean(shy) - 1.0E0 # Set Mean around 0.0
shy_detrend = shy_tmp/np.std(shy_tmp) # Set standard deviation to 1.0
cm = common_mode / np.mean(common_mode) - 1.0E0
cm_detrend = cm/np.std(cm)
fig, axs = plt.subplots()
plt.plot(shx_detrend, label='X-possition')
plt.plot(shy_detrend, label='Y-possition')
plt.xlabel('Image Sequence Number')
plt.ylabel('Relative Detector Possition')
plt.title('Time-series Detrending Vectors')
axs.xaxis.set_major_locator(ticker.MultipleLocator(1000))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(100))
axs.yaxis.set_major_locator(ticker.MultipleLocator(0.5))
axs.yaxis.set_minor_locator(ticker.MultipleLocator(0.1))
plt.legend()
plt.show()
Create arrays of the vectors used for detrending.#
From Sing et al. 2019: Systematic errors are often removed by a parameterized deterministic model, where the non-transit photometric trends are found to correlate with a number \(n\) of external parameters (or optical state vectors, \(X\)). These parameters describe changes in the instrument or other external factors as a function of time during the observations, and are fit with a coefficient for each optical state parameter, \(p_n\), to model and remove (or detrend) the photometric light curves.
When including systematic trends, the total parameterized model of the flux measurements over time, \(f(t)\), can be modeled as a combination of the theoretical transit model, \(T(t,\theta)\) (which depends upon the transit parameters \(\theta\)), the total baseline flux detected from the star, \(F_0\), and the systematics error model \(S(x)\) giving,
\(f(t) = T(t,\theta)\times F_0 \times S(x)\).
We will use a linear model for the instrument systematic effects.
\(S(x)= p_1 x + p_2 y + p_3 x^2 + p_4 y^2 + p_5 x y + p_6 cm + p_7 \phi \)
\(cm\) is the common_mode trend, and \(\phi\) is a linear time trend which helps remove changing H\(_2\)O ice within the H\(_2\)O spectral feature.
shx = shx_detrend
shy = shy_detrend
common_mode = cm_detrend
XX = np.array([shx, shy, shx**2, shy**2, shx*shy, common_mode, np.ones(number_of_images)]) # Detrending array without linear time trend
XX = np.transpose(XX)
XXX = np.array([shx, shy, shx**2, shy**2, shx*shy, common_mode, phase, np.ones(number_of_images)]) # Detrending array with with linear time trend
XXX = np.transpose(XXX)
Linear Regression can be used to quickly determine the parameters \(p_n\) using the out-of-transit data.
Here, we take a wavelength bin of the data (pixels 170 to 200) to make a time series. The out-of-transit points are selected and a linear regression of \(S(x)\) is done to determine the optical state parameters \(p_n\)
pix1 = 170 # wavelength bin lower range
pix2 = 200 # wavelength bin upper range
y = np.sum(all_spec_1D[pix1:pix2, :], axis=0) # flux over a selected wavelength bin
msize = plt.rcParams['lines.markersize'] ** 2. # default marker size
plt.rcParams['figure.figsize'] = [10.0, 3.0] # Figure dimensions
fig, axs = plt.subplots()
f = plt.plot(wsdata_all, all_spec_1D[:, 0], linewidth=2, zorder=0) # Plot Region of wavelength bin
plt.fill_between(wsdata_all[pix1:pix2], 0, all_spec_1D[pix1:pix2, 0], alpha=0.5)
plt.xlabel(r'Wavelength ($\mu$m)')
plt.ylabel('Flux (e-)')
plt.title('1D Extracted Spectrum')
axs.xaxis.set_major_locator(ticker.MultipleLocator(0.5))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(0.1))
plt.annotate('H$_2$O', xy=(3.0, 42000))
plt.annotate('CO$_2$', xy=(4.2, 42000))
plt.show()
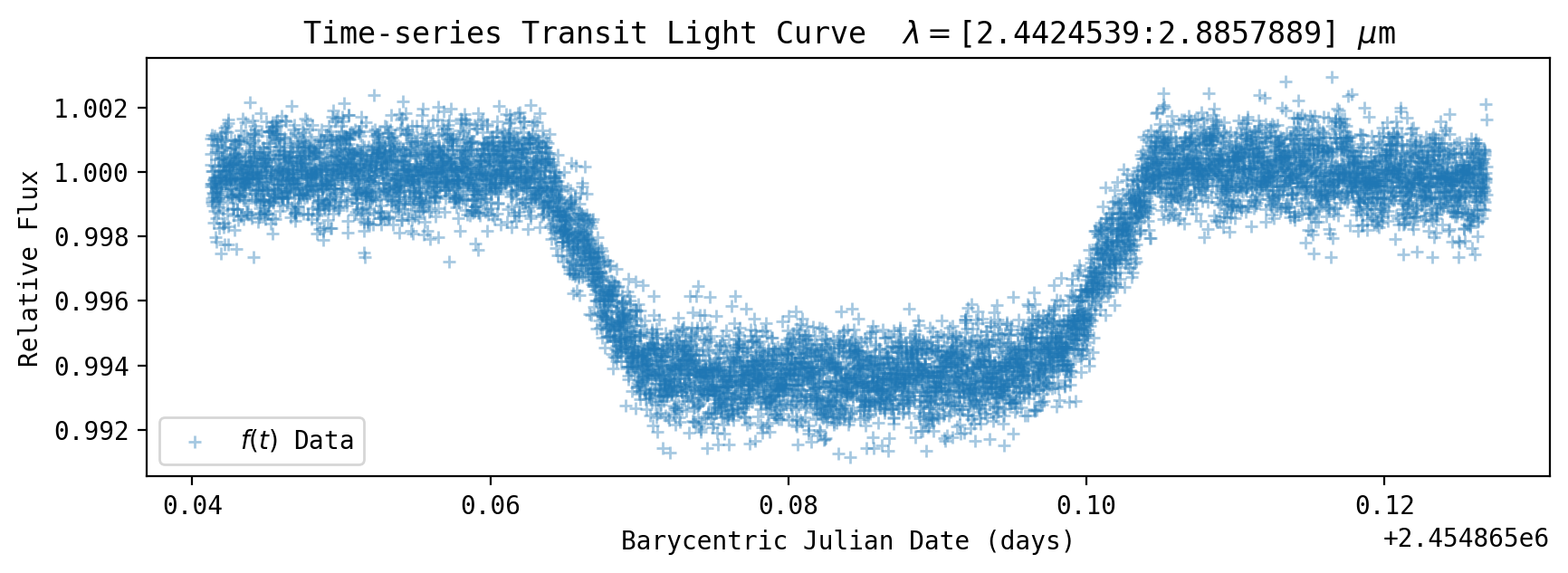
fig, axs = plt.subplots()
plt.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.4, marker='+', edgecolors='blue')
plt.xlabel('Barycentric Julian Date (days)')
plt.ylabel('Relative Flux')
plt.title(r'Time-series Transit Light Curve $\lambda=$['+str(wsdata_all[pix1])+':'+str(wsdata_all[pix2]) + r'] $\mu$m')
plt.legend()
plt.show()
regressor = LinearRegression()
regressor.fit(XX[outtransit], y[outtransit]/np.mean(y[outtransit]))
print('Linear Regression Coefficients:')
print(regressor.coef_)
/tmp/ipykernel_1956/1502773371.py:21: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.4, marker='+', edgecolors='blue')
Linear Regression Coefficients:
[ 2.35589645e-04 2.33200386e-04 -1.43527088e-04 -4.65560797e-05
1.94065907e-04 -4.51855007e-04 0.00000000e+00]
The coefficients are on the order of ~10\(^{-4}\) so the trends have an amplitude on the order of 100’s of ppm.
Visualize the fit
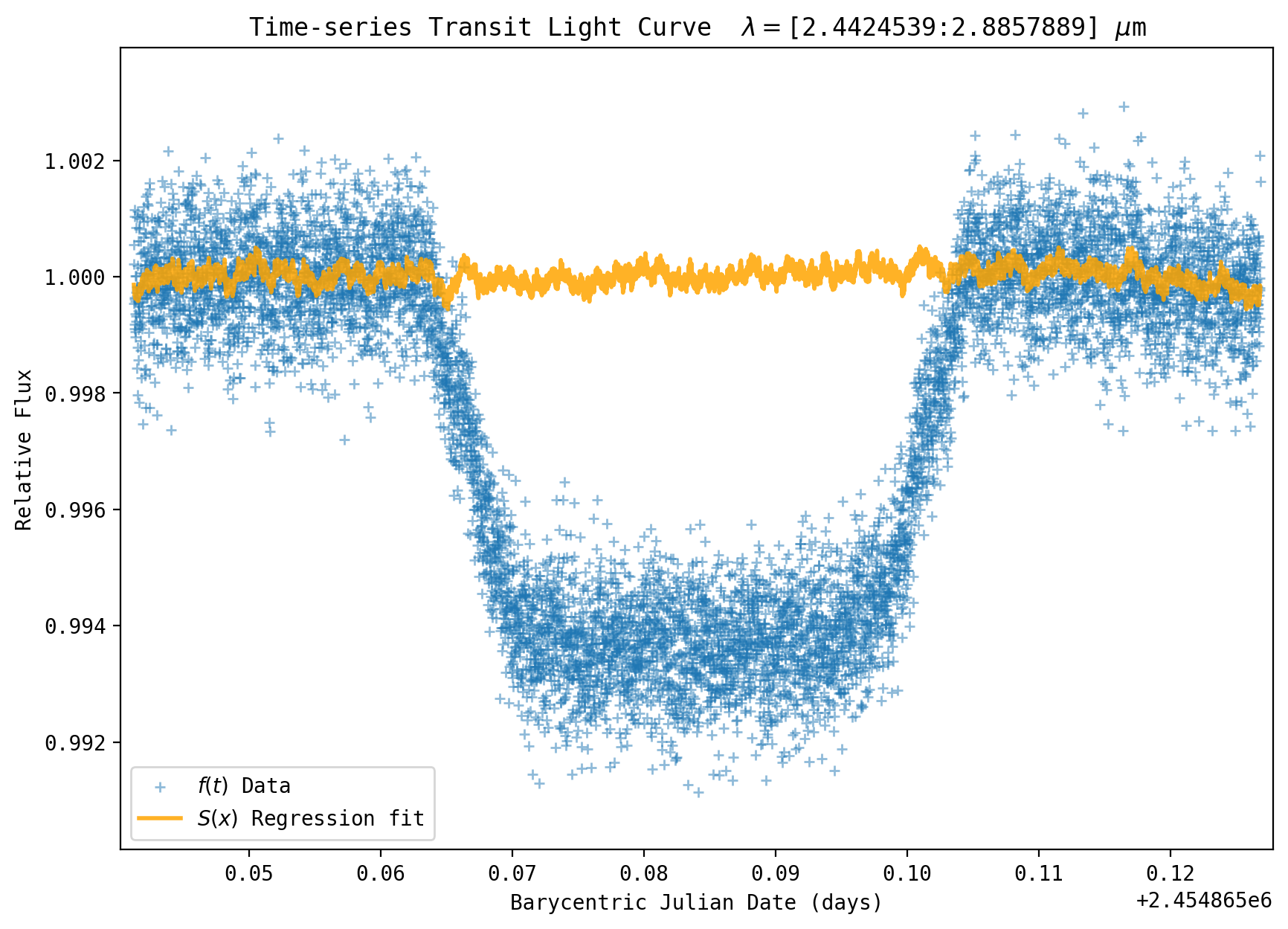
yfit = regressor.predict(XX) # Project the fit over the whole time series
plt.rcParams['figure.figsize'] = [10.0, 7.0] # Figure dimensions
msize = plt.rcParams['lines.markersize'] ** 2. # default marker size
plt.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
f = plt.plot(bjd, yfit, label='$S(x)$ Regression fit ', linewidth=2, color='orange', zorder=2, alpha=0.85)
plt.xlabel('Barycentric Julian Date (days)')
plt.ylabel('Relative Flux')
plt.title(r'Time-series Transit Light Curve $\lambda=$['+str(wsdata_all[pix1])+':'+str(wsdata_all[pix2]) + r'] $\mu$m')
axs.xaxis.set_major_locator(ticker.MultipleLocator(0.01))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(0.005))
axs.yaxis.set_major_locator(ticker.MultipleLocator(0.002))
axs.yaxis.set_minor_locator(ticker.MultipleLocator(0.001))
yplot = y / np.mean(y[outtransit])
plt.ylim(yplot.min() * 0.999, yplot.max()*1.001)
plt.xlim(bjd.min()-0.001, bjd.max()+0.001)
plt.legend(loc='lower left')
plt.show()
/tmp/ipykernel_1956/3911644354.py:5: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
Transit and Limb-Darkening Model Functions#
Define a functions used by the fitting routines. These which will take the transit and systematic parameters and create our full transit light curve model
\(model = T(t,\theta)\times F_0 \times S(x)\)
compares it to the data
\(y = f(t)\)
by returning the residuals
\((y-model)/(\sigma_y)\)
To calculate the transit model, here we use Mandel and Agol (2002) as coded in python by H. Wakeford (ExoTiC-ISM).
To calculate the stellar limb-darkening, we use the procedure from Sing et al. (2010) which uses stellar models and fits for non-linear limb darkening coefficients, with a module as coded in python by H. Wakeford (ExoTiC-ISM).
A new orbit is first calculated based on the system parameters of \(a/R_{star}\), the cosine of the inclination \(cos(i)\), and the orbital phase \(\phi\). The inputs are the orbit distance between the planet-star center \(b\) at each phase, limb-darkening parameters (\(c_1,c_2,c_3,c_4\)), and the planet-to-star radius ratio \(R_p/R_{star}\).
@custom_model
def nonlinear_limb_darkening(x, c0=0.0, c1=0.0, c2=0.0, c3=0.0):
"""
Define non-linear limb darkening model with four parameters c0, c1, c2, c3.
"""
model = (1. - (c0 * (1. - x ** (1. / 2)) + c1 * (1. - x ** (2. / 2)) + c2 * (1. - x ** (3. / 2)) + c3 *
(1. - x ** (4. / 2))))
return model
@custom_model
def quadratic_limb_darkening(x, aLD=0.0, bLD=0.0):
"""
Define linear limb darkening model with parameters aLD and bLD.
"""
model = 1. - aLD * (1. - x) - bLD * (1. - x) ** (4. / 2.)
return model
def limb_dark_fit(grating, wsdata, M_H, Teff, logg, dirsen, ld_model='1D'):
"""
Calculates stellar limb-darkening coefficients for a given wavelength bin.
Currently supports:
HST STIS G750L, G750M, G430L gratings
HST WFC3 UVIS/G280, IR/G102, IR/G141 grisms
What is used for 1D models - Kurucz (?)
Procedure from Sing et al. (2010, A&A, 510, A21).
Uses 3D limb darkening from Magic et al. (2015, A&A, 573, 90).
Uses photon FLUX Sum over (lambda*dlamba).
:param grating: string; grating to use ('G430L','G750L','G750M', 'G280', 'G102', 'G141')
:param wsdata: array; data wavelength solution
:param M_H: float; stellar metallicity
:param Teff: float; stellar effective temperature (K)
:param logg: float; stellar gravity
:param dirsen: string; path to main limb darkening directory
:param ld_model: string; '1D' or '3D', makes choice between limb darkening models; default is 1D
:return: uLD: float; linear limb darkening coefficient
aLD, bLD: float; quadratic limb darkening coefficients
cp1, cp2, cp3, cp4: float; three-parameter limb darkening coefficients
c1, c2, c3, c4: float; non-linear limb-darkening coefficients
"""
print('You are using the', str(ld_model), 'limb darkening models.')
if ld_model == '1D':
direc = os.path.join(dirsen, 'Kurucz')
print('Current Directories Entered:')
print(' ' + dirsen)
print(' ' + direc)
# Select metallicity
M_H_Grid = np.array([-0.1, -0.2, -0.3, -0.5, -1.0, -1.5, -2.0, -2.5, -3.0, -3.5, -4.0, -4.5, -5.0, 0.0, 0.1, 0.2, 0.3, 0.5, 1.0])
M_H_Grid_load = np.array([0, 1, 2, 3, 5, 7, 8, 9, 10, 11, 12, 13, 14, 17, 20, 21, 22, 23, 24])
optM = (abs(M_H - M_H_Grid)).argmin()
MH_ind = M_H_Grid_load[optM]
# Determine which model is to be used, by using the input metallicity M_H to figure out the file name we need
direc = 'Kurucz'
file_list = 'kuruczlist.sav'
sav1 = readsav(os.path.join(dirsen, file_list))
model = bytes.decode(sav1['li'][MH_ind]) # Convert object of type "byte" to "string"
# Select Teff and subsequently logg
Teff_Grid = np.array([3500, 3750, 4000, 4250, 4500, 4750, 5000, 5250, 5500, 5750, 6000, 6250, 6500])
optT = (abs(Teff - Teff_Grid)).argmin()
logg_Grid = np.array([4.0, 4.5, 5.0])
optG = (abs(logg - logg_Grid)).argmin()
if logg_Grid[optG] == 4.0:
Teff_Grid_load = np.array([8, 19, 30, 41, 52, 63, 74, 85, 96, 107, 118, 129, 138])
elif logg_Grid[optG] == 4.5:
Teff_Grid_load = np.array([9, 20, 31, 42, 53, 64, 75, 86, 97, 108, 119, 129, 139])
elif logg_Grid[optG] == 5.0:
Teff_Grid_load = np.array([10, 21, 32, 43, 54, 65, 76, 87, 98, 109, 120, 130, 140])
# Where in the model file is the section for the Teff we want? Index T_ind tells us that.
T_ind = Teff_Grid_load[optT]
header_rows = 3 # How many rows in each section we ignore for the data reading
data_rows = 1221 # How many rows of data we read
line_skip_data = (T_ind + 1) * header_rows + T_ind * data_rows # Calculate how many lines in the model file we need to skip in order to get to the part we need (for the Teff we want).
line_skip_header = T_ind * (data_rows + header_rows)
# Read the header, in case we want to have the actual Teff, logg and M_H info.
# headerinfo is a pandas object.
headerinfo = pd.read_csv(os.path.join(dirsen, direc, model), delim_whitespace=True, header=None,
skiprows=line_skip_header, nrows=1)
Teff_model = headerinfo[1].values[0]
logg_model = headerinfo[3].values[0]
MH_model = headerinfo[6].values[0]
MH_model = float(MH_model[1:-1])
print('\nClosest values to your inputs:')
print('Teff: ', Teff_model)
print('M_H: ', MH_model)
print('log(g): ', logg_model)
# Read the data; data is a pandas object.
data = pd.read_csv(os.path.join(dirsen, direc, model), delim_whitespace=True, header=None,
skiprows=line_skip_data, nrows=data_rows)
# Unpack the data
ws = data[0].values * 10 # Import wavelength data
f0 = data[1].values / (ws * ws)
f1 = data[2].values * f0 / 100000.
f2 = data[3].values * f0 / 100000.
f3 = data[4].values * f0 / 100000.
f4 = data[5].values * f0 / 100000.
f5 = data[6].values * f0 / 100000.
f6 = data[7].values * f0 / 100000.
f7 = data[8].values * f0 / 100000.
f8 = data[9].values * f0 / 100000.
f9 = data[10].values * f0 / 100000.
f10 = data[11].values * f0 / 100000.
f11 = data[12].values * f0 / 100000.
f12 = data[13].values * f0 / 100000.
f13 = data[14].values * f0 / 100000.
f14 = data[15].values * f0 / 100000.
f15 = data[16].values * f0 / 100000.
f16 = data[17].values * f0 / 100000.
# Make single big array of them
fcalc = np.array([f0, f1, f2, f3, f4, f5, f6, f7, f8, f9, f10, f11, f12, f13, f14, f15, f16])
phot1 = np.zeros(fcalc.shape[0])
# Define mu
mu = np.array([1.000, .900, .800, .700, .600, .500, .400, .300, .250, .200, .150, .125, .100, .075, .050, .025, .010])
# Passed on to main body of function are: ws, fcalc, phot1, mu
elif ld_model == '3D':
direc = os.path.join(dirsen, '3DGrid')
print('Current Directories Entered:')
print(' ' + dirsen)
print(' ' + direc)
# Select metallicity
M_H_Grid = np.array([-3.0, -2.0, -1.0, 0.0]) # Available metallicity values in 3D models
M_H_Grid_load = ['30', '20', '10', '00'] # The according identifiers to individual available M_H values
optM = (abs(M_H - M_H_Grid)).argmin() # Find index at which the closes M_H values from available values is to the input M_H.
# Select Teff
Teff_Grid = np.array([4000, 4500, 5000, 5500, 5777, 6000, 6500, 7000]) # Available Teff values in 3D models
optT = (abs(Teff - Teff_Grid)).argmin() # Find index at which the Teff values is, that is closest to input Teff.
# Select logg, depending on Teff. If several logg possibilities are given for one Teff, pick the one that is
# closest to user input (logg).
if Teff_Grid[optT] == 4000:
logg_Grid = np.array([1.5, 2.0, 2.5])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 4500:
logg_Grid = np.array([2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 5000:
logg_Grid = np.array([2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 5500:
logg_Grid = np.array([3.0, 3.5, 4.0, 4.5, 5.0])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 5777:
logg_Grid = np.array([4.4])
optG = 0
elif Teff_Grid[optT] == 6000:
logg_Grid = np.array([3.5, 4.0, 4.5])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 6500:
logg_Grid = np.array([4.0, 4.5])
optG = (abs(logg - logg_Grid)).argmin()
elif Teff_Grid[optT] == 7000:
logg_Grid = np.array([4.5])
optG = 0
# Select Teff and Log g. Mtxt, Ttxt and Gtxt are then put together as string to load correct files.
Mtxt = M_H_Grid_load[optM]
Ttxt = "{:2.0f}".format(Teff_Grid[optT] / 100)
if Teff_Grid[optT] == 5777:
Ttxt = "{:4.0f}".format(Teff_Grid[optT])
Gtxt = "{:2.0f}".format(logg_Grid[optG] * 10)
#
file = 'mmu_t' + Ttxt + 'g' + Gtxt + 'm' + Mtxt + 'v05.flx'
print('Filename:', file)
# Read data from IDL .sav file
sav = readsav(os.path.join(direc, file)) # readsav reads an IDL .sav file
ws = sav['mmd'].lam[0] # read in wavelength
flux = sav['mmd'].flx # read in flux
Teff_model = Teff_Grid[optT]
logg_model = logg_Grid[optG]
MH_model = str(M_H_Grid[optM])
print('\nClosest values to your inputs:')
print('Teff : ', Teff_model)
print('M_H : ', MH_model)
print('log(g): ', logg_model)
f0 = flux[0]
f1 = flux[1]
f2 = flux[2]
f3 = flux[3]
f4 = flux[4]
f5 = flux[5]
f6 = flux[6]
f7 = flux[7]
f8 = flux[8]
f9 = flux[9]
f10 = flux[10]
# Make single big array of them
fcalc = np.array([f0, f1, f2, f3, f4, f5, f6, f7, f8, f9, f10])
phot1 = np.zeros(fcalc.shape[0])
# Mu from grid
# 0.00000 0.0100000 0.0500000 0.100000 0.200000 0.300000 0.500000 0.700000 0.800000 0.900000 1.00000
mu = sav['mmd'].mu
# Passed on to main body of function are: ws, fcalc, phot1, mu
# Load response function and interpolate onto kurucz model grid
# FOR STIS
if grating == 'G430L':
sav = readsav(os.path.join(dirsen, 'G430L.STIS.sensitivity.sav')) # wssens,sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 3
if grating == 'G750M':
sav = readsav(os.path.join(dirsen, 'G750M.STIS.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 0.554
if grating == 'G750L':
sav = readsav(os.path.join(dirsen, 'G750L.STIS.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 4.882
# FOR WFC3
if grating == 'G141': # http://www.stsci.edu/hst/acs/analysis/reference_files/synphot_tables.html
sav = readsav(os.path.join(dirsen, 'G141.WFC3.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 1
if grating == 'G102': # http://www.stsci.edu/hst/acs/analysis/reference_files/synphot_tables.html
sav = readsav(os.path.join(dirsen, 'G141.WFC3.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 1
if grating == 'G280': # http://www.stsci.edu/hst/acs/analysis/reference_files/synphot_tables.html
sav = readsav(os.path.join(dirsen, 'G280.WFC3.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 1
# FOR JWST
if grating == 'NIRSpecPrism': # http://www.stsci.edu/hst/acs/analysis/reference_files/synphot_tables.html
sav = readsav(os.path.join(dirsen, 'NIRSpec.prism.sensitivity.sav')) # wssens, sensitivity
wssens = sav['wssens']
sensitivity = sav['sensitivity']
wdel = 12
widek = np.arange(len(wsdata))
wsHST = wssens
wsHST = np.concatenate((np.array([wsHST[0] - wdel - wdel, wsHST[0] - wdel]),
wsHST,
np.array([wsHST[len(wsHST) - 1] + wdel,
wsHST[len(wsHST) - 1] + wdel + wdel])))
respoutHST = sensitivity / np.max(sensitivity)
respoutHST = np.concatenate((np.zeros(2), respoutHST, np.zeros(2)))
inter_resp = interp1d(wsHST, respoutHST, bounds_error=False, fill_value=0)
respout = inter_resp(ws) # interpolate sensitivity curve onto model wavelength grid
wsdata = np.concatenate((np.array([wsdata[0] - wdel - wdel, wsdata[0] - wdel]), wsdata,
np.array([wsdata[len(wsdata) - 1] + wdel, wsdata[len(wsdata) - 1] + wdel + wdel])))
respwavebin = wsdata / wsdata * 0.0
widek = widek + 2 # need to add two indicies to compensate for padding with 2 zeros
respwavebin[widek] = 1.0
data_resp = interp1d(wsdata, respwavebin, bounds_error=False, fill_value=0)
reswavebinout = data_resp(ws) # interpolate data onto model wavelength grid
# Integrate over the spectra to make synthetic photometric points.
for i in range(fcalc.shape[0]): # Loop over spectra at diff angles
fcal = fcalc[i, :]
Tot = int_tabulated(ws, ws * respout * reswavebinout)
phot1[i] = (int_tabulated(ws, ws * respout * reswavebinout * fcal, sort=True)) / Tot
if ld_model == '1D':
yall = phot1 / phot1[0]
elif ld_model == '3D':
yall = phot1 / phot1[10]
# Co = np.zeros((6, 4)) # NOT-REUSED
# A = [0.0, 0.0, 0.0, 0.0] # c1, c2, c3, c4 # NOT-REUSED
x = mu[1:] # wavelength
y = yall[1:] # flux
# weights = x / x # NOT-REUSED
# Start fitting the different models
fitter = LevMarLSQFitter()
# Fit a four parameter non-linear limb darkening model and get fitted variables, c1, c2, c3, c4.
corot_4_param = nonlinear_limb_darkening()
corot_4_param = fitter(corot_4_param, x, y)
c1, c2, c3, c4 = corot_4_param.parameters
# Fit a three parameter non-linear limb darkening model and get fitted variables, cp2, cp3, cp4 (cp1 = 0).
corot_3_param = nonlinear_limb_darkening()
corot_3_param.c0.fixed = True # 3 param is just 4 param with c0 = 0.0
corot_3_param = fitter(corot_3_param, x, y)
cp1, cp2, cp3, cp4 = corot_3_param.parameters
# Fit a quadratic limb darkening model and get fitted parameters aLD and bLD.
quadratic = quadratic_limb_darkening()
quadratic = fitter(quadratic, x, y)
aLD, bLD = quadratic.parameters
# Fit a linear limb darkening model and get fitted variable uLD.
linear = nonlinear_limb_darkening()
linear.c0.fixed = True
linear.c2.fixed = True
linear.c3.fixed = True
linear = fitter(linear, x, y)
uLD = linear.c1.value
print('\nLimb darkening parameters:')
print("4param \t{:0.8f}\t{:0.8f}\t{:0.8f}\t{:0.8f}".format(c1, c2, c3, c4))
print("3param \t{:0.8f}\t{:0.8f}\t{:0.8f}".format(cp2, cp3, cp4))
print("Quad \t{:0.8f}\t{:0.8f}".format(aLD, bLD))
print("Linear \t{:0.8f}".format(uLD))
return uLD, c1, c2, c3, c4, cp1, cp2, cp3, cp4, aLD, bLD
def int_tabulated(X, F, sort=False):
Xsegments = len(X) - 1
# Sort vectors into ascending order.
if not sort:
ii = np.argsort(X)
X = X[ii]
F = F[ii]
while (Xsegments % 4) != 0:
Xsegments = Xsegments + 1
Xmin = np.min(X)
Xmax = np.max(X)
# Uniform step size.
h = (Xmax + 0.0 - Xmin) / Xsegments
# Compute the interpolates at Xgrid.
# x values of interpolates >> Xgrid = h * FINDGEN(Xsegments + 1L) + Xmin
z = splev(h * np.arange(Xsegments + 1) + Xmin, splrep(X, F))
# Compute the integral using the 5-point Newton-Cotes formula.
ii = (np.arange((len(z) - 1) / 4, dtype=int) + 1) * 4
return np.sum(2.0 * h * (7.0 * (z[ii - 4] + z[ii]) + 32.0 * (z[ii - 3] + z[ii - 1]) + 12.0 * z[ii - 2]) / 45.0)
Now define the transit model function#
def occultnl(rl, c1, c2, c3, c4, b0):
"""
MANDEL & AGOL (2002) transit model.
:param rl: float, transit depth (Rp/R*)
:param c1: float, limb darkening parameter 1
:param c2: float, limb darkening parameter 2
:param c3: float, limb darkening parameter 3
:param c4: float, limb darkening parameter 4
:param b0: impact parameter in stellar radii
:return: mulimb0: limb-darkened transit model, mulimbf: lightcurves for each component that you put in the model
"""
mulimb0 = occultuniform(b0, rl)
bt0 = b0
fac = np.max(np.abs(mulimb0 - 1))
if fac == 0:
fac = 1e-6 # DKS edit
omega = 4 * ((1 - c1 - c2 - c3 - c4) / 4 + c1 / 5 + c2 / 6 + c3 / 7 + c4 / 8)
nb = len(b0)
indx = np.where(mulimb0 != 1.0)[0]
if len(indx) == 0:
indx = -1
mulimb = mulimb0[indx]
mulimbf = np.zeros((5, nb))
mulimbf[0, :] = mulimbf[0, :] + 1.
mulimbf[1, :] = mulimbf[1, :] + 0.8
mulimbf[2, :] = mulimbf[2, :] + 2 / 3
mulimbf[3, :] = mulimbf[3, :] + 4 / 7
mulimbf[4, :] = mulimbf[4, :] + 0.5
nr = np.int64(2)
dmumax = 1.0
while (dmumax > fac * 1.e-3) and (nr <= 131072):
# print(nr)
mulimbp = mulimb
nr = nr * 2
dt = 0.5 * np.pi / nr
t = dt * np.arange(nr + 1)
th = t + 0.5 * dt
r = np.sin(t)
sig = np.sqrt(np.cos(th[nr - 1]))
mulimbhalf = sig ** 3 * mulimb0[indx] / (1 - r[nr - 1])
mulimb1 = sig ** 4 * mulimb0[indx] / (1 - r[nr - 1])
mulimb3half = sig ** 5 * mulimb0[indx] / (1 - r[nr - 1])
mulimb2 = sig ** 6 * mulimb0[indx] / (1 - r[nr - 1])
for i in range(1, nr):
mu = occultuniform(b0[indx] / r[i], rl / r[i])
sig1 = np.sqrt(np.cos(th[i - 1]))
sig2 = np.sqrt(np.cos(th[i]))
mulimbhalf = mulimbhalf + r[i] ** 2 * mu * (sig1 ** 3 / (r[i] - r[i - 1]) - sig2 ** 3 / (r[i + 1] - r[i]))
mulimb1 = mulimb1 + r[i] ** 2 * mu * (sig1 ** 4 / (r[i] - r[i - 1]) - sig2 ** 4 / (r[i + 1] - r[i]))
mulimb3half = mulimb3half + r[i] ** 2 * mu * (sig1 ** 5 / (r[i] - r[i - 1]) - sig2 ** 5 / (r[i + 1] - r[i]))
mulimb2 = mulimb2 + r[i] ** 2 * mu * (sig1 ** 6 / (r[i] - r[i - 1]) - sig2 ** 6 / (r[i + 1] - r[i]))
mulimb = ((1 - c1 - c2 - c3 - c4) * mulimb0[
indx] + c1 * mulimbhalf * dt + c2 * mulimb1 * dt + c3 * mulimb3half * dt + c4 * mulimb2 * dt) / omega
ix1 = np.where(mulimb + mulimbp != 0.)[0]
if len(ix1) == 0:
ix1 = -1
# print(ix1)
# python cannot index on single values so you need to use atlest_1d for the below to work when mulimb is a single value
dmumax = np.max(np.abs(np.atleast_1d(mulimb)[ix1] - np.atleast_1d(mulimbp)[ix1]) / (
np.atleast_1d(mulimb)[ix1] + np.atleast_1d(mulimbp)[ix1]))
mulimbf[0, indx] = np.atleast_1d(mulimb0)[indx]
mulimbf[1, indx] = mulimbhalf * dt
mulimbf[2, indx] = mulimb1 * dt
mulimbf[3, indx] = mulimb3half * dt
mulimbf[4, indx] = mulimb2 * dt
np.atleast_1d(mulimb0)[indx] = mulimb
b0 = bt0
return mulimb0, mulimbf
def occultuniform(b0, w):
"""
Compute the lightcurve for occultation of a uniform source without microlensing (Mandel & Agol 2002).
:param b0: array; impact parameter in units of stellar radii
:param w: array; occulting star size in units of stellar radius
:return: muo1: float; fraction of flux at each b0 for a uniform source
"""
if np.abs(w - 0.5) < 1.0e-3:
w = 0.5
nb = len(np.atleast_1d(b0))
muo1 = np.zeros(nb)
for i in range(nb):
# substitute z=b0(i) to shorten expressions
z = np.atleast_1d(b0)[i]
# z = z.value # stripping it of astropy units
if z >= 1+w:
muo1[i] = 1.0
continue
if w >= 1 and z <= w-1:
muo1[i] = 0.0
continue
if z >= np.abs(1-w) and z <= 1+w:
kap1 = np.arccos(np.min(np.append((1 - w ** 2 + z ** 2) / 2 / z, 1.)))
kap0 = np.arccos(np.min(np.append((w ** 2 + z ** 2 - 1) / 2 / w / z, 1.)))
lambdae = w ** 2 * kap0 + kap1
lambdae = (lambdae - 0.5 * np.sqrt(np.max(np.append(4. * z ** 2 - (1 + z ** 2 - w ** 2) ** 2, 0.)))) / np.pi
muo1[i] = 1 - lambdae
if z <= 1-w:
muo1[i] = 1 - w ** 2
continue
return muo1
Now define the function to generate the transit light curve and compare it to the data#
# Functions to call and calculate models
def residual(p, phase, x, y, err, c1, c2, c3, c4):
# calculate new orbit
b0 = p['a_Rs'].value * np.sqrt((np.sin(phase * 2 * np.pi)) ** 2 + (p['cosinc'].value * np.cos(phase * 2 * np.pi)) ** 2)
# Select indicies between first and fourth contact
intransit = (b0-p['rprs'].value < 1.0E0).nonzero()
# Make light curve model, set all values initially to 1.0
light_curve = b0/b0
mulimb0, mulimbf = occultnl(p['rprs'].value, c1, c2, c3, c4, b0[intransit]) # Madel and Agol
light_curve[intransit] = mulimb0
model = (light_curve) * p['f0'].value * (p['Fslope'].value * phase + p['xsh'].value * shx + p['x2sh'].value * shx**2. + p['ysh'].value * shy + p['y2sh'].value * shy**2. + p['xysh'].value * shy * shx + p['comm'].value * common_mode + 1.0) # transit model is baseline flux X transit model X systematics model
chi2now = np.sum((y-model)**2/err**2)
res = np.std((y-model)/p['f0'].value)
print("rprs: ", p['rprs'].value, "current chi^2=", chi2now, ' scatter ', res, end="\r")
return (y-model)/err
# return np.sum((y-model)**2/err**2)
A function is also defined to return just the transit model \(T(t,\theta)\)
def model_fine(p): # Make Transit model with a fine grid for plotting purposes
b0 = p['a_Rs'].value * np.sqrt((np.sin(phase_fine * 2 * np.pi)) ** 2 + (p['cosinc'].value * np.cos(phase_fine * 2 * np.pi)) ** 2)
mulimb0, mulimbf = occultnl(p['rprs'].value, c1, c2, c3, c4, b0) # Madel and Agol
model_fine = mulimb0
return model_fine
Now add a transit model to the Example Light curve. Here, we’ve compute the limb darkening coefficients, then use them in the transit light curve.
wave1 = wsdata_all[pix1]
wave2 = wsdata_all[pix2]
bin_wave_index = ((wsdata_all > wave1) & (wsdata_all <= wave2)).nonzero()
wsdata = wsdata_all[bin_wave_index]*1E4 # Select wavelength bin values (um=> angstroms)
_uLD, c1, c2, c3, c4, _cp1, _cp2, _cp3, _cp4, aLD, bLD = limb_dark_fit(grating, wsdata, M_H, Teff, logg, limb_dark_directory, ld_model)
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.87682090 -0.73755228 0.57723174 -0.19560435
3param 2.19231542 -3.11195060 1.36536769
Quad 0.02781785 0.37167814
Linear 0.34303525
Now run the transit model.
The transit parameters such as inclination and \(a/R_{star}\) have been setup at the beginning of the notebook.
# Run the Transit Model
rl = 0.0825 # Planet-to-star Radius Ratio
b0 = a_Rs * np.sqrt((np.sin(phase * 2 * np.pi)) ** 2 + (np.cos(inc) * np.cos(phase * 2 * np.pi)) ** 2)
intransit = (b0-rl < 1.0E0).nonzero() # Select indicies between first and fourth contact
mulimb0, mulimbf = occultnl(rl, c1, c2, c3, c4, b0) # Mandel & Agol non-linear limb darkened transit model
model = mulimb0*yfit
# plot
plt.rcParams['figure.figsize'] = [10.0, 7.0] # Figure dimensions
msize = plt.rcParams['lines.markersize'] ** 2. # default marker size
fig = plt.figure(constrained_layout=True)
gs = fig.add_gridspec(3, 1, hspace=0.00, wspace=0.00)
ax1 = fig.add_subplot(gs[0:2, :])
ax1.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
ax1.plot(bjd, model, label='$S(x)$ Regression fit ', linewidth=2, color='orange', zorder=2, alpha=0.85)
ax1.xaxis.set_ticklabels([])
plt.ylabel('Relative Flux')
plt.title(r'Time-series Transit Light Curve $\lambda=$['+str(wsdata_all[pix1])+':'+str(wsdata_all[pix2]) + r'] $\mu$m')
ax1.xaxis.set_major_locator(ticker.MultipleLocator(0.01))
ax1.xaxis.set_minor_locator(ticker.MultipleLocator(0.005))
ax1.yaxis.set_major_locator(ticker.MultipleLocator(0.002))
ax1.yaxis.set_minor_locator(ticker.MultipleLocator(0.001))
yplot = y/np.mean(y[outtransit])
plt.ylim(yplot.min()*0.999, yplot.max()*1.001)
plt.xlim(bjd.min()-0.001, bjd.max()+0.001)
plt.legend()
fig.add_subplot(ax1)
# Residual
ax2 = fig.add_subplot(gs[2, :])
ax2.scatter(bjd, 1E6*(y/np.mean(y[outtransit])-model), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
wsb, wsb_bin_edges, binnumber = stats.binned_statistic(bjd, 1E6*(y/np.mean(y[outtransit])-model), bins=256)
plt.scatter(wsb_bin_edges[1:], wsb, linewidth=2, alpha=0.75, facecolors='orange', edgecolors='none', marker='o', zorder=25)
plt.xlabel('Barycentric Julian Date (days)')
plt.ylabel('Residual (ppm)')
ax2.xaxis.set_major_locator(ticker.MultipleLocator(0.01))
ax2.xaxis.set_minor_locator(ticker.MultipleLocator(0.005))
yplot = y / np.mean(y[outtransit])
plt.xlim(bjd.min()-0.001, bjd.max()+0.001)
fig.add_subplot(ax2)
plt.show()
# print chi^2 value
err = np.sqrt(y) / np.mean(y[outtransit])
print('Chi^2 = '+str(np.sum((y/np.mean(y[outtransit])-model)**2/err**2)))
print('Residual Standard Deviation : '+str(1E6*np.std((y/np.mean(y[outtransit])-model)))+' ppm')
print('256 Bin Standard Deviation :'+str(np.std(wsb))+' ppm')
/tmp/ipykernel_1956/1695522111.py:16: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(bjd, y/np.mean(y[outtransit]), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/1695522111.py:32: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(bjd, 1E6*(y/np.mean(y[outtransit])-model), label='$f(t)$ Data', zorder=1, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue')
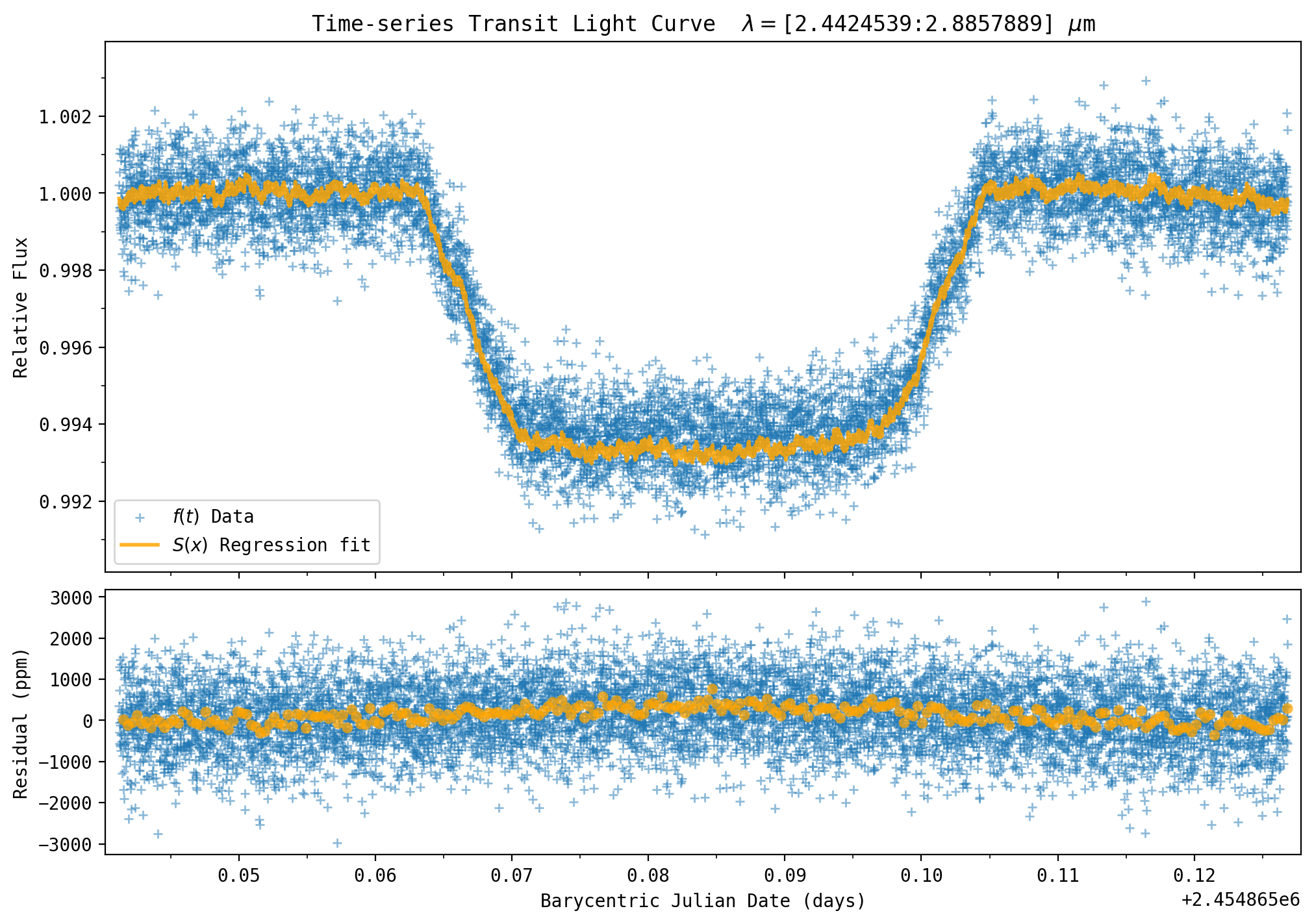
Chi^2 = 9265.443817708492
Residual Standard Deviation : 789.8093583096888 ppm
256 Bin Standard Deviation :196.83309935696437 ppm
Note that the model transit depth is a little too deep compared to the data. The planet radius needs to be smaller, and the parameter \(rl\) is closer to 0.08. As an exercise you can re-run the above cell changing the planet radius to \(rl\)=0.0805 and compare the \(\chi^2\) value to the previous default value (\(\chi^2\)=9265.4 at \(rl\) = 0.0825).
FIT Transit Light Curves#
Now we can fit each light curve, optimizing the fit parameters with a least-squares fit. Here a Levenberg-Marquart fit is used to find a \(\chi^2\) minimum and estimate uncertainties using the lmfit package (https://lmfit.github.io/lmfit-py/fitting.html).
In practice, first we would fit the white light curve, which consists of summing over all of the wavelengths in the entire 1D spectra. This can then be used to fit for the system parameters such as inclination and transit time, and then the spectroscopic channels are then fixed to these values as they are wavelength-independent. However, the CV3 data required the overall variations of the lamp to be removed, which prevents our use of using this data for a white light curve analysis. Here, we proceed to fitting for the spectroscopic light curve bins.
The steps are as follows:
1) Wavelength Bin is selected
2) Limb-darkening coefficients are calculated from a stellar model for each bin.
3) An initial linear regression is performed on the out-of-transit data to start the
systematic fit parameters, this greatly speeds up the fit as those parameters start
near their global minimum.
4) The fit is started, and some statistics are output during the minimization
5) Once the best-fit is found, a number of statistics are displayed
6) Finally, several plots are generated which are stored as PDFs and the next bin is started.
These steps are performed for each spectral bin.
In this example, the planet radius is set to vary in the fit along with the baseline flux and instrument systematic parameters.
Setup Wavelengths to fit over#
The spectra must be binned in wavelength to get sufficient counts to reach ~100 ppm levels needed. The spectra has significant counts from about pixel 100 to 400, we start at pixel \(k0\) and bin the spectra by \(wk\) pixels.
Several arrays are also defined.
k0 = 113 # 98 #100
kend = 392 # 422
wk = 15
number_of_bins = int((kend-k0)/wk)
wsd = np.zeros((number_of_bins))
werr = np.zeros((number_of_bins))
rprs = np.zeros((number_of_bins))
rerr = np.zeros((number_of_bins))
sig_r = np.zeros((number_of_bins))
sig_w = np.zeros((number_of_bins))
beta = np.zeros((number_of_bins))
depth = np.zeros((number_of_bins))
depth_err = np.zeros((number_of_bins))
Loop Over Wavelength Bins Fitting Each Lightcurve#
Note this step takes considerable time to complete (~20 min, few minutes/bin)#
Each wavelength bin is fit for the transit+systematics model. Various outputs including plots are saved. Can skip to the next cells to load pre-computed results.
k = k0 # wavelength to start
# --------------------------------------------------------------------------
# Loop over wavelength bins and fit for each one
for bin in range(0, number_of_bins):
# Select wavelength bin
wave1 = wsdata_all[k]
wave2 = wsdata_all[k+wk]
# Indicies to select for wavelgth bin
bin_wave_index = ((wsdata_all > wave1) & (wsdata_all <= wave2)).nonzero()
# make light curve bin
wave_bin_counts = np.sum(all_spec_1D[k+1:k+wk, :], axis=0) # Sum Wavelength pixels
wave_bin_counts_err = np.sqrt(wave_bin_counts) # adopt photon noise for errors
# Calculate Limb Darkening
wsdata = wsdata_all[bin_wave_index]*1E4 # Select wavelength bin values (um=> angstroms)
_uLD, c1, c2, c3, c4, _cp1, _cp2, _cp3, _cp4, aLD, bLD = limb_dark_fit(grating, wsdata, M_H, Teff, logg, limb_dark_directory, ld_model)
print('\nc1 = {}'.format(c1))
print('c2 = {}'.format(c2))
print('c3 = {}'.format(c3))
print('c4 = {}'.format(c4))
print('')
# u = [c1,c2,c3,c4] # limb darkening coefficients
# u = [aLD, bLD]
# Make initial model
# Setup LMFIT
x = bjd # X data
y = wave_bin_counts # Y data
err = wave_bin_counts_err # Y Error
# Perform Quick Linear regression on out-of-transit data to obtain accurate starting Detector fit values
if wave1 > 2.7 and wave1 < 3.45:
regressor.fit(XXX[outtransit], y[outtransit]/np.mean(y[outtransit]))
else:
regressor.fit(XX[outtransit], y[outtransit]/np.mean(y[outtransit]))
# create a set of Parameters for LMFIT https://lmfit.github.io/lmfit-py/parameters.html
# class Parameter(name, value=None, vary=True, min=- inf, max=inf, expr=None, brute_step=None, user_data=None)¶
# Set vary=0 to fix
# Set vary=1 to fit
p = lmfit.Parameters() # object to store L-M fit Parameters # Parameter Name
p.add('cosinc', value=np.cos(inc), vary=0) # inclination, vary cos(inclin)
p.add('rho_star', value=rho_star, vary=0) # stellar density
p.add('a_Rs', value=a_Rs, vary=0) # a/Rstar
p.add('rprs', value=rp, vary=1, min=0, max=1) # planet-to-star radius ratio
p.add('t0', value=t0, vary=0) # Transit T0
p.add('f0', value=np.mean(y[outtransit]), vary=1, min=0) # Baseline Flux
p.add('ecc', value=ecc, vary=0, min=0, max=1) # eccentricity
p.add('omega', value=omega, vary=0) # arguments of periatron
# Turn on a linear slope in water feature to account for presumably changing H2O ice builtup on widow during cryogenic test
if wave1 > 2.7 and wave1 < 3.45:
p.add('Fslope', value=regressor.coef_[6], vary=1) # Orbital phase
else:
p.add('Fslope', value=0, vary=0) # Orbital phase
p.add('xsh', value=regressor.coef_[0], vary=1) # Detector X-shift detrending
p.add('ysh', value=regressor.coef_[1], vary=1) # Detector X-shift detrending
p.add('x2sh', value=regressor.coef_[2], vary=1) # Detector X^2-shift detrending
p.add('y2sh', value=regressor.coef_[3], vary=1) # Detector Y^2-shift detrending
p.add('xysh', value=regressor.coef_[4], vary=1) # Detector X*Y detrending
p.add('comm', value=regressor.coef_[5], vary=1) # Common-Mode detrending
# Perform Minimization https://lmfit.github.io/lmfit-py/fitting.html
# create Minimizer
# mini = lmfit.Minimizer(residual, p, nan_policy='omit',fcn_args=(phase,x,y,err)
print('')
print('Fitting Bin', bin, ' Wavelength =', np.mean(wsdata)/1E4, ' Range= [', wave1, ':', wave2, ']')
# solve with Levenberg-Marquardt using the
result = lmfit.minimize(residual, params=p, args=(phase, x, y, err, c1, c2, c3, c4))
# result = mini.minimize(method='emcee')
print('')
print('Re-Fitting Bin', bin, ' Wavelength =', np.mean(wsdata)/1E4, ' Range= [', wave1, ':', wave2, ']')
# --------------------------------------------------------------------------
print("")
print("redchi", result.redchi)
print("chi2", result.chisqr)
print("nfree", result.nfree)
print("bic", result.bic)
print("aic", result.aic)
print("L-M FIT Variable")
print(lmfit.fit_report(result.params))
text_file = open(save_directory+'JWST_NIRSpec_Prism_fit_light_curve_bin'+str(bin)+'_statistics.txt', "w")
n = text_file.write("\nredchi "+str(result.redchi))
n = text_file.write("\nchi2 "+str(result.chisqr))
n = text_file.write("\nnfree "+str(result.nfree))
n = text_file.write("\nbic "+str(result.bic))
n = text_file.write("\naic "+str(result.aic))
n = text_file.write(lmfit.fit_report(result.params))
# file-output.py
# Update with best-fit parameters
p['rho_star'].value = result.params['rho_star'].value
p['cosinc'].value = result.params['cosinc'].value
p['rprs'].value = result.params['rprs'].value
p['t0'].value = result.params['t0'].value
p['f0'].value = result.params['f0'].value
p['Fslope'].value = result.params['Fslope'].value
p['xsh'].value = result.params['xsh'].value
p['ysh'].value = result.params['ysh'].value
p['x2sh'].value = result.params['x2sh'].value
p['y2sh'].value = result.params['y2sh'].value
p['xysh'].value = result.params['xysh'].value
p['comm'].value = result.params['comm'].value
# Update Fit Spectra arrays
wsd[bin] = np.mean(wsdata)/1E4
werr[bin] = (wsdata.max()-wsdata.min())/2E4
rprs[bin] = result.params['rprs'].value
rerr[bin] = result.params['rprs'].stderr
# Calculate Bestfit Model
final_model = y-result.residual*err
final_model_fine = model_fine(p)
# More Stats
resid = (y-final_model)/p['f0'].value
residppm = 1E6*(y-final_model)/p['f0'].value
residerr = err/p['f0'].value
sigma = np.std((y-final_model)/p['f0'].value)*1E6
print("Residual standard deviation (ppm) : ", 1E6*np.std((y-final_model)/p['f0'].value))
print("Photon noise (ppm) : ", (1/np.sqrt(p['f0'].value))*1E6)
print("Photon noise performance (%) : ", (1/np.sqrt(p['f0'].value))*1E6 / (sigma) * 100)
n = text_file.write("\nResidual standard deviation (ppm) : "+str(1E6*np.std((y-final_model)/p['f0'].value)))
n = text_file.write("\nPhoton noise (ppm) : "+str((1/np.sqrt(p['f0'].value))*1E6))
n = text_file.write("\nPhoton noise performance (%) : "+str((1/np.sqrt(p['f0'].value))*1E6 / (sigma) * 100))
# Measure Rednoise with Binning Technique
sig0 = np.std(resid)
bins = number_of_images / binmeasure
wsb, wsb_bin_edges, binnumber = stats.binned_statistic(bjd, resid, bins=bins)
sig_binned = np.std(wsb)
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
if np.isnan(sigrednoise):
sigrednoise = 0 # if no rednoise detected, set to zero
sigwhite = np.sqrt(sig0**2-sigrednoise**2)
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
if np.isnan(sigrednoise):
sigrednoise = 0 # if no rednoise detected, set to zero
beta[bin] = np.sqrt(sig0**2+binmeasure*sigrednoise**2)/sig0
print("White noise (ppm) : ", 1E6*sigwhite)
print("Red noise (ppm) : ", 1E6*sigrednoise)
print("Transit depth measured error (ppm) : ", 2E6*result.params['rprs'].value*result.params['rprs'].stderr)
n = text_file.write("\nWhite noise (ppm) : "+str(1E6*sigwhite))
n = text_file.write("\nRed noise (ppm) : "+str(1E6*sigrednoise))
n = text_file.write("\nTransit depth measured error (ppm) : "+str(2E6*result.params['rprs'].value*result.params['rprs'].stderr))
text_file.close()
depth[bin] = 1E6*result.params['rprs'].value**2
depth_err[bin] = 2E6*result.params['rprs'].value*result.params['rprs'].stderr
sig_r[bin] = sigrednoise*1E6
sig_w[bin] = sigwhite*1E6
# --------------------------------------------------------------------------
# ---------------------------------------------------------
# Write Fit Spectra to ascii file
ascii_data = Table([wsd, werr, rprs, rerr, depth, depth_err, sig_w, sig_r, beta], names=['Wavelength Center (um)', 'Wavelength half-width (um)', 'Rp/Rs', 'Rp/Rs 1-sigma error', 'Transit Depth (ppm)', 'Transit Depth error', 'Sigma_white (ppm)', 'Sigma_red (ppm)', 'Beta Rednoise Inflation factor'])
ascii.write(ascii_data, save_directory+'JWST_NIRSpec_Prism_fit_transmission_spectra.csv', format='csv', overwrite=True)
# ---------------------------------------------------------
msize = plt.rcParams['lines.markersize'] ** 2. # default marker size
# Plot data models
# plot
plt.rcParams['figure.figsize'] = [10.0, 7.0] # Figure dimensions
msize = plt.rcParams['lines.markersize'] ** 2. # default marker size
fig = plt.figure(constrained_layout=True)
gs = fig.add_gridspec(3, 1, hspace=0.00, wspace=0.00)
ax1 = fig.add_subplot(gs[0:2, :])
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
ax1.plot(x, final_model/p['f0'].value, linewidth=1, color='orange', alpha=0.8, zorder=15) # overplot Transit model at data
ax1.xaxis.set_ticklabels([])
plt.ylabel('Relative Flux')
plt.title(r'Time-series Transit Light Curve $\lambda=$['+str(wave1)+':'+str(wave2) + r'] $\mu$m')
ax1.xaxis.set_major_locator(ticker.MultipleLocator(0.01))
ax1.xaxis.set_minor_locator(ticker.MultipleLocator(0.005))
ax1.yaxis.set_major_locator(ticker.MultipleLocator(0.002))
ax1.yaxis.set_minor_locator(ticker.MultipleLocator(0.001))
yplot = y/np.mean(y[outtransit])
plt.ylim(yplot.min()*0.999, yplot.max()*1.001)
plt.xlim(bjd.min()-0.001, bjd.max()+0.001)
fig.add_subplot(ax1)
# Residual
ax2 = fig.add_subplot(gs[2, :])
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
wsb, wsb_bin_edges, binnumber = stats.binned_statistic(bjd, residppm, bins=256)
plt.scatter(wsb_bin_edges[1:], wsb, linewidth=2, alpha=0.75, facecolors='orange', edgecolors='none', marker='o', zorder=25)
plt.xlabel('Barycentric Julian Date (days)')
plt.ylabel('Residual (ppm)')
plt.plot([bjd.min(), bjd.max()], [0, 0], color='black', zorder=10)
plt.plot([bjd.min(), bjd.max()], [sigma, sigma], linestyle='--', color='black', zorder=15)
plt.plot([bjd.min(), bjd.max()], [-sigma, -sigma], linestyle='--', color='black', zorder=20)
ax2.xaxis.set_major_locator(ticker.MultipleLocator(0.01))
ax2.xaxis.set_minor_locator(ticker.MultipleLocator(0.005))
yplot = y/np.mean(y[outtransit])
plt.xlim(bjd.min()-0.001, bjd.max()+0.001)
fig.add_subplot(ax2)
# save
pp = PdfPages(save_directory+'JWST_NIRSpec_Prism_fit_light_curve_bin'+str(bin)+'_lightcurve.pdf')
plt.savefig(pp, format='pdf')
pp.close()
plt.clf()
# plot systematic corrected light curve
b0 = p['a_Rs'].value * np.sqrt((np.sin(phase * 2 * np.pi)) ** 2 + (p['cosinc'].value * np.cos(phase * 2 * np.pi)) ** 2)
intransit = (b0-p['rprs'].value < 1.0E0).nonzero()
light_curve = b0/b0
mulimb0, mulimbf = occultnl(p['rprs'].value, c1, c2, c3, c4, b0[intransit]) # Madel and Agol
light_curve[intransit] = mulimb0
fig, axs = plt.subplots()
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
plt.xlabel('BJD')
plt.ylabel('Relative Flux')
plt.plot(x, light_curve, linewidth=2, color='orange', alpha=0.8, zorder=15) # overplot Transit model at data
pp = PdfPages(save_directory+'JWST_NIRSpec_Prism_fit_light_curve_bin'+str(bin)+'_corrected.pdf')
plt.savefig(pp, format='pdf')
pp.close()
plt.clf()
plt.close('all') # close all figures
# --------------------------------------------------------------------------
k = k + wk # step wavelength index to next bin
print('** Can Now View Output PDFs in ', save_directory)
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.93487505 -0.53100613 0.43228652 -0.16656941
3param 2.59284758 -3.50115591 1.49775430
Quad 0.08088643 0.45654030
Linear 0.46807479
c1 = 0.9348750506946152
c2 = -0.531006129789577
c3 = 0.43228651640755095
c4 = -0.1665694117104222
Fitting Bin 0 Wavelength = 1.5502650866666667 Range= [ 1.3906398 : 1.6900786 ]
rprs: 0.07869514581104897 current chi^2= 13085.417569552985 scatter 0.0023922892879259575
Re-Fitting Bin 0 Wavelength = 1.5502650866666667 Range= [ 1.3906398 : 1.6900786 ]
redchi 1.5989024400724565
chi2 13085.417569552985
nfree 8184
bic 3908.7316936329157
aic 3852.644386854681
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.07869515 +/- 5.4580e-04 (0.69%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 278433.709 +/- 22.2711416 (0.01%) (init = 278410.6)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -0.00112032 +/- 9.9569e-05 (8.89%) (init = -0.001091413)
ysh: 3.3601e-05 +/- 9.2623e-05 (275.66%) (init = 0.0002380423)
x2sh: -4.1703e-04 +/- 2.2616e-04 (54.23%) (init = -0.0003157139)
y2sh: -3.3390e-04 +/- 2.2424e-04 (67.16%) (init = -0.0001744718)
xysh: 6.7719e-04 +/- 4.4533e-04 (65.76%) (init = 0.0003395803)
comm: 5.4547e-04 +/- 8.2561e-05 (15.14%) (init = 0.0002697908)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9755
C(x2sh, y2sh) = +0.9376
C(rprs, f0) = +0.8090
C(xsh, ysh) = -0.6325
C(xsh, comm) = -0.4878
C(f0, x2sh) = -0.3857
C(f0, y2sh) = -0.3280
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2739
C(xsh, x2sh) = +0.2427
C(xsh, xysh) = -0.2134
C(rprs, x2sh) = -0.1917
C(xysh, comm) = +0.1795
C(xsh, y2sh) = +0.1757
C(x2sh, comm) = -0.1724
C(rprs, xsh) = -0.1644
C(y2sh, comm) = -0.1486
C(rprs, y2sh) = -0.1369
C(rprs, ysh) = +0.1341
C(f0, xsh) = -0.1185
Residual standard deviation (ppm) : 2392.2892879259575
Photon noise (ppm) : 1895.1303791648108
Photon noise performance (%) : 79.21827802054122
White noise (ppm) : 2387.129444269294
Red noise (ppm) : 157.34479866780455
Transit depth measured error (ppm) : 85.90311947299497
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 1.12080976 -0.97843713 0.75312858 -0.25673953
3param 2.76671119 -3.96262532 1.73859692
Quad 0.01052393 0.47328324
Linear 0.41191184
c1 = 1.1208097603789067
c2 = -0.9784371279899322
c3 = 0.7531285754201472
c4 = -0.25673952710770237
Fitting Bin 1 Wavelength = 1.84545072 Range= [ 1.6900786 : 1.9783379 ]
rprs: 0.08047594288462401 current chi^2= 14107.324261909878 scatter 0.0015893597541472527
Re-Fitting Bin 1 Wavelength = 1.84545072 Range= [ 1.6900786 : 1.9783379 ]
redchi 1.7237688492069743
chi2 14107.324261909878
nfree 8184
bic 4524.734590843977
aic 4468.6472840657425
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08047594 +/- 3.5164e-04 (0.44%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 679862.651 +/- 36.2424305 (0.01%) (init = 679718.7)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -0.00119414 +/- 6.6177e-05 (5.54%) (init = -0.001210365)
ysh: -5.0144e-04 +/- 6.1567e-05 (12.28%) (init = -0.0004319937)
x2sh: -1.1190e-04 +/- 1.5033e-04 (134.34%) (init = -8.991835e-05)
y2sh: -6.4121e-05 +/- 1.4905e-04 (232.45%) (init = -1.47937e-05)
xysh: -3.4284e-05 +/- 2.9598e-04 (863.31%) (init = -0.0001337487)
comm: 0.00107817 +/- 5.4864e-05 (5.09%) (init = 0.001005529)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9755
C(x2sh, y2sh) = +0.9376
C(rprs, f0) = +0.8102
C(xsh, ysh) = -0.6328
C(xsh, comm) = -0.4875
C(f0, x2sh) = -0.3863
C(f0, y2sh) = -0.3290
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2747
C(xsh, x2sh) = +0.2431
C(xsh, xysh) = -0.2139
C(rprs, x2sh) = -0.1930
C(xysh, comm) = +0.1791
C(xsh, y2sh) = +0.1762
C(x2sh, comm) = -0.1719
C(rprs, xsh) = -0.1654
C(y2sh, comm) = -0.1482
C(rprs, y2sh) = -0.1388
C(rprs, ysh) = +0.1364
C(f0, xsh) = -0.1196
Residual standard deviation (ppm) : 1589.3597541472527
Photon noise (ppm) : 1212.8006148401325
Photon noise performance (%) : 76.3074949944761
White noise (ppm) : 1588.7050679155338
Red noise (ppm) : 45.70298419235961
Transit depth measured error (ppm) : 56.59740034216333
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 1.05423283 -0.97018497 0.77456621 -0.26658145
3param 2.55249877 -3.66106843 1.61023054
Quad 0.01394080 0.43332492
Linear 0.38144036
c1 = 1.0542328323187433
c2 = -0.9701849696940038
c3 = 0.7745662054908382
c4 = -0.2665814455066517
Fitting Bin 2 Wavelength = 2.1223476199999998 Range= [ 1.9783379 : 2.2448517 ]
rprs: 0.07999433245668902 current chi^2= 12525.313078681707 scatter 0.0012094149112427549
Re-Fitting Bin 2 Wavelength = 2.1223476199999998 Range= [ 1.9783379 : 2.2448517 ]
redchi 1.530463474912232
chi2 12525.313078681706
nfree 8184
bic 3550.3578704200436
aic 3494.270563641809
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.07999433 +/- 2.6809e-04 (0.34%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 1042975.58 +/- 42.3147415 (0.00%) (init = 1043280)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -7.4025e-04 +/- 5.0367e-05 (6.80%) (init = -0.0006767627)
ysh: -3.6016e-04 +/- 4.6857e-05 (13.01%) (init = -0.0003522163)
x2sh: 1.4337e-04 +/- 1.1440e-04 (79.80%) (init = 0.0001534885)
y2sh: 3.2762e-04 +/- 1.1343e-04 (34.62%) (init = 0.0003162653)
xysh: -3.3426e-04 +/- 2.2524e-04 (67.38%) (init = -0.0003427445)
comm: 7.9760e-04 +/- 4.1747e-05 (5.23%) (init = 0.0007132)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9755
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8104
C(xsh, ysh) = -0.6329
C(xsh, comm) = -0.4874
C(f0, x2sh) = -0.3866
C(f0, y2sh) = -0.3294
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2751
C(xsh, x2sh) = +0.2435
C(xsh, xysh) = -0.2142
C(rprs, x2sh) = -0.1935
C(xysh, comm) = +0.1793
C(xsh, y2sh) = +0.1765
C(x2sh, comm) = -0.1721
C(rprs, xsh) = -0.1669
C(y2sh, comm) = -0.1484
C(rprs, y2sh) = -0.1393
C(rprs, ysh) = +0.1378
C(f0, xsh) = -0.1209
C(rprs, xysh) = +0.1004
Residual standard deviation (ppm) : 1209.414911242755
Photon noise (ppm) : 979.1808909666712
Photon noise performance (%) : 80.96318987505265
White noise (ppm) : 1206.4904195622446
Red noise (ppm) : 84.21931993071638
Transit depth measured error (ppm) : 42.890979153541636
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.93455579 -0.84286559 0.70782403 -0.25114311
3param 2.27992131 -3.22427512 1.41261223
Quad 0.03322149 0.38713152
Linear 0.36154478
c1 = 0.9345557878924728
c2 = -0.842865591448989
c3 = 0.7078240285892669
c4 = -0.25114311341177076
Fitting Bin 3 Wavelength = 2.377201606666667 Range= [ 2.2448517 : 2.4898652 ]
rprs: 0.08152564128527812 current chi^2= 10199.771177809664 scatter 0.001054037346435932
Re-Fitting Bin 3 Wavelength = 2.377201606666667 Range= [ 2.2448517 : 2.4898652 ]
redchi 1.2463063511497634
chi2 10199.771177809664
nfree 8184
bic 1867.832838218284
aic 1811.7455314400497
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08152564 +/- 2.2958e-04 (0.28%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 1117867.74 +/- 39.5908739 (0.00%) (init = 1117799)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -5.0800e-04 +/- 4.3896e-05 (8.64%) (init = -0.0004562807)
ysh: 1.2772e-04 +/- 4.0840e-05 (31.98%) (init = 0.000158932)
x2sh: 1.9435e-04 +/- 9.9708e-05 (51.30%) (init = 0.0001891058)
y2sh: 1.5157e-04 +/- 9.8860e-05 (65.22%) (init = 0.000155217)
xysh: -4.0988e-04 +/- 1.9630e-04 (47.89%) (init = -0.0003957601)
comm: 1.8396e-04 +/- 3.6386e-05 (19.78%) (init = 0.0001085827)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9819
C(x2sh, xysh) = -0.9755
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8110
C(xsh, ysh) = -0.6329
C(xsh, comm) = -0.4873
C(f0, x2sh) = -0.3865
C(f0, y2sh) = -0.3296
C(ysh, comm) = -0.3012
C(f0, xysh) = +0.2751
C(xsh, x2sh) = +0.2435
C(xsh, xysh) = -0.2141
C(rprs, x2sh) = -0.1937
C(xysh, comm) = +0.1792
C(xsh, y2sh) = +0.1765
C(x2sh, comm) = -0.1720
C(rprs, xsh) = -0.1664
C(y2sh, comm) = -0.1483
C(rprs, y2sh) = -0.1399
C(rprs, ysh) = +0.1377
C(f0, xsh) = -0.1207
C(rprs, xysh) = +0.1007
Residual standard deviation (ppm) : 1054.037346435932
Photon noise (ppm) : 945.8119322073638
Photon noise performance (%) : 89.73229794992406
White noise (ppm) : 1052.8206317440454
Red noise (ppm) : 50.72926682911007
Transit depth measured error (ppm) : 37.434013904412495
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.87680367 -0.73477190 0.58280906 -0.20005969
3param 2.19503822 -3.10630077 1.36088167
Quad 0.03045602 0.37301051
Linear 0.34680339
c1 = 0.8768036657165874
c2 = -0.7347718979202172
c3 = 0.5828090572725704
c4 = -0.2000596941008768
Fitting Bin 4 Wavelength = 2.6120002933333333 Range= [ 2.4898652 : 2.7162005 ]
rprs: 0.08079801924266616 current chi^2= 9205.709924232755 scatter 0.0010482768870580229
Re-Fitting Bin 4 Wavelength = 2.6120002933333333 Range= [ 2.4898652 : 2.7162005 ]
redchi 1.1248423661085964
chi2 9205.709924232753
nfree 8184
bic 1027.8140906872184
aic 971.7267839089841
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08079802 +/- 2.2966e-04 (0.28%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 1020091.41 +/- 35.9154006 (0.00%) (init = 1020040)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 4.9600e-04 +/- 4.3663e-05 (8.80%) (init = 0.0004393402)
ysh: 2.2241e-04 +/- 4.0620e-05 (18.26%) (init = 0.000309657)
x2sh: -2.9802e-04 +/- 9.9151e-05 (33.27%) (init = -0.0003434394)
y2sh: -2.1077e-04 +/- 9.8311e-05 (46.64%) (init = -0.0002199046)
xysh: 4.9765e-04 +/- 1.9521e-04 (39.23%) (init = 0.0005561113)
comm: -6.8653e-04 +/- 3.6185e-05 (5.27%) (init = -0.0007121339)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9755
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8109
C(xsh, ysh) = -0.6329
C(xsh, comm) = -0.4873
C(f0, x2sh) = -0.3866
C(f0, y2sh) = -0.3297
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2753
C(xsh, x2sh) = +0.2441
C(xsh, xysh) = -0.2145
C(rprs, x2sh) = -0.1938
C(xysh, comm) = +0.1795
C(xsh, y2sh) = +0.1769
C(x2sh, comm) = -0.1724
C(rprs, xsh) = -0.1678
C(y2sh, comm) = -0.1486
C(rprs, y2sh) = -0.1399
C(rprs, ysh) = +0.1386
C(f0, xsh) = -0.1220
C(rprs, xysh) = +0.1009
Residual standard deviation (ppm) : 1048.2768870580228
Photon noise (ppm) : 990.1031808272959
Photon noise performance (%) : 94.45054002917199
White noise (ppm) : 1047.7390803816916
Red noise (ppm) : 33.64007590200573
Transit depth measured error (ppm) : 37.1121852245308
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.86128574 -0.71348923 0.53201916 -0.17329851
3param 2.16446825 -3.09179973 1.36001686
Quad 0.02096721 0.36578104
Linear 0.33118333
c1 = 0.8612857385139974
c2 = -0.713489233845713
c3 = 0.5320191611044409
c4 = -0.17329850590331394
Fitting Bin 5 Wavelength = 2.82970062 Range= [ 2.7162005 : 2.9267646 ]
rprs: 0.0798569649594219 current chi^2= 9962.93384572772 scatter 0.0016130786363043458
Re-Fitting Bin 5 Wavelength = 2.82970062 Range= [ 2.7162005 : 2.9267646 ]
redchi 1.2175160510482368
chi2 9962.93384572772
nfree 8183
bic 1684.383398479687
aic 1621.2851783541735
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.07985696 +/- 4.4930e-04 (0.56%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 466305.818 +/- 27.4132622 (0.01%) (init = 466320.8)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: -0.03200602 +/- 0.00907390 (28.35%) (init = -0.03885871)
xsh: 5.9845e-04 +/- 7.7661e-05 (12.98%) (init = 0.0005208206)
ysh: 2.0512e-04 +/- 6.4429e-05 (31.41%) (init = 0.0001880271)
x2sh: 1.3205e-04 +/- 1.5407e-04 (116.67%) (init = 0.0003234828)
y2sh: 1.2008e-04 +/- 1.5150e-04 (126.17%) (init = 0.0003067711)
xysh: -1.9185e-04 +/- 3.0269e-04 (157.77%) (init = -0.0005659834)
comm: -4.1856e-04 +/- 6.2462e-05 (14.92%) (init = -0.0002515501)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9796
C(x2sh, xysh) = -0.9758
C(x2sh, y2sh) = +0.9346
C(rprs, f0) = +0.8292
C(rprs, Fslope) = -0.6095
C(Fslope, xsh) = -0.5011
C(Fslope, comm) = -0.4531
C(xsh, ysh) = -0.4105
C(f0, Fslope) = -0.3889
C(f0, x2sh) = -0.2996
C(f0, y2sh) = -0.2832
C(xsh, x2sh) = +0.2785
C(rprs, comm) = +0.2590
C(rprs, ysh) = +0.2547
C(xsh, xysh) = -0.2457
C(Fslope, ysh) = -0.2417
C(f0, xysh) = +0.2044
C(rprs, xsh) = +0.1894
C(xsh, y2sh) = +0.1791
C(ysh, comm) = -0.1509
C(xsh, comm) = -0.1489
C(f0, ysh) = +0.1401
C(Fslope, x2sh) = -0.1377
C(f0, comm) = +0.1331
C(Fslope, xysh) = +0.1223
C(y2sh, comm) = -0.1088
C(xysh, comm) = +0.1034
Residual standard deviation (ppm) : 1613.0786363043458
Photon noise (ppm) : 1464.4163972762306
Photon noise performance (%) : 90.7839434679571
White noise (ppm) : 1609.088864053264
Red noise (ppm) : 113.60427859881536
Transit depth measured error (ppm) : 71.75961566001148
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.83139630 -0.69648917 0.51398704 -0.16463361
3param 2.08159374 -2.98407343 1.31547068
Quad 0.01845846 0.35019596
Linear 0.31545699
c1 = 0.8313962953828634
c2 = -0.69648916712715
c3 = 0.5139870433675813
c4 = -0.1646336112166497
Fitting Bin 6 Wavelength = 3.03297832 Range= [ 2.9267646 : 3.1240093 ]
rprs: 0.07996217817640366 current chi^2= 9377.921035738047 scatter 0.0016759321966837164
Re-Fitting Bin 6 Wavelength = 3.03297832 Range= [ 2.9267646 : 3.1240093 ]
redchi 1.1460248118951541
chi2 9377.921035738047
nfree 8183
bic 1188.6568837001298
aic 1125.5586635746163
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.07996218 +/- 4.6570e-04 (0.58%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 406639.550 +/- 24.8600565 (0.01%) (init = 406639.5)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: -0.02768365 +/- 0.00943268 (34.07%) (init = -0.0245558)
xsh: 9.5268e-05 +/- 8.0679e-05 (84.69%) (init = 0.0001198049)
ysh: -2.8172e-04 +/- 6.6954e-05 (23.77%) (init = -0.0003972918)
x2sh: 1.2084e-04 +/- 1.6007e-04 (132.46%) (init = 0.0001082623)
y2sh: 8.5765e-05 +/- 1.5741e-04 (183.53%) (init = 4.592033e-05)
xysh: -2.0445e-04 +/- 3.1449e-04 (153.82%) (init = -0.0001742409)
comm: 3.1334e-04 +/- 6.4895e-05 (20.71%) (init = 0.0003524477)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9796
C(x2sh, xysh) = -0.9758
C(x2sh, y2sh) = +0.9347
C(rprs, f0) = +0.8296
C(rprs, Fslope) = -0.6101
C(Fslope, xsh) = -0.5009
C(Fslope, comm) = -0.4532
C(xsh, ysh) = -0.4102
C(f0, Fslope) = -0.3898
C(f0, x2sh) = -0.2995
C(f0, y2sh) = -0.2834
C(xsh, x2sh) = +0.2783
C(rprs, comm) = +0.2591
C(rprs, ysh) = +0.2558
C(xsh, xysh) = -0.2456
C(Fslope, ysh) = -0.2424
C(f0, xysh) = +0.2044
C(rprs, xsh) = +0.1896
C(xsh, y2sh) = +0.1790
C(ysh, comm) = -0.1505
C(xsh, comm) = -0.1488
C(f0, ysh) = +0.1414
C(Fslope, x2sh) = -0.1374
C(f0, comm) = +0.1332
C(Fslope, xysh) = +0.1221
C(y2sh, comm) = -0.1088
C(xysh, comm) = +0.1033
Residual standard deviation (ppm) : 1675.9321966837165
Photon noise (ppm) : 1568.1774042089141
Photon noise performance (%) : 93.57045632943718
White noise (ppm) : 1673.2041442027448
Red noise (ppm) : 95.77217562834234
Transit depth measured error (ppm) : 74.47713547007491
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.81389247 -0.73763493 0.57400093 -0.18855617
3param 1.98195953 -2.85041302 1.26038669
Quad 0.01890988 0.33067264
Linear 0.29935083
c1 = 0.8138924681638973
c2 = -0.7376349257280954
c3 = 0.5740009254129774
c4 = -0.18855617472606742
Fitting Bin 7 Wavelength = 3.2240248533333333 Range= [ 3.1240093 : 3.3099063 ]
rprs: 0.08173193067865869 current chi^2= 9016.09530809757 scatter 0.0015709991669581843
Re-Fitting Bin 7 Wavelength = 3.2240248533333333 Range= [ 3.1240093 : 3.3099063 ]
redchi 1.101808054270753
chi2 9016.09530809757
nfree 8183
bic 866.3282848080157
aic 803.2300646825022
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08173193 +/- 4.2849e-04 (0.52%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 444771.966 +/- 25.5781982 (0.01%) (init = 444786.4)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: -0.04678146 +/- 0.00886101 (18.94%) (init = -0.04274347)
xsh: -2.0829e-04 +/- 7.5679e-05 (36.33%) (init = -0.0001220049)
ysh: -6.4652e-04 +/- 6.2784e-05 (9.71%) (init = -0.000715279)
x2sh: -2.1765e-04 +/- 1.5007e-04 (68.95%) (init = -9.505015e-05)
y2sh: -1.9149e-04 +/- 1.4759e-04 (77.07%) (init = -9.269764e-05)
xysh: 4.4090e-04 +/- 2.9486e-04 (66.88%) (init = 0.0002427406)
comm: 9.2486e-04 +/- 6.0847e-05 (6.58%) (init = 0.0008865038)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9796
C(x2sh, xysh) = -0.9759
C(x2sh, y2sh) = +0.9346
C(rprs, f0) = +0.8308
C(rprs, Fslope) = -0.6122
C(Fslope, xsh) = -0.5019
C(Fslope, comm) = -0.4531
C(xsh, ysh) = -0.4091
C(f0, Fslope) = -0.3929
C(f0, x2sh) = -0.2986
C(f0, y2sh) = -0.2833
C(xsh, x2sh) = +0.2779
C(rprs, comm) = +0.2589
C(rprs, ysh) = +0.2566
C(xsh, xysh) = -0.2455
C(Fslope, ysh) = -0.2432
C(f0, xysh) = +0.2037
C(rprs, xsh) = +0.1922
C(xsh, y2sh) = +0.1785
C(ysh, comm) = -0.1502
C(xsh, comm) = -0.1481
C(f0, ysh) = +0.1427
C(Fslope, x2sh) = -0.1371
C(f0, comm) = +0.1336
C(Fslope, xysh) = +0.1219
C(y2sh, comm) = -0.1088
C(xysh, comm) = +0.1032
Residual standard deviation (ppm) : 1570.9991669581843
Photon noise (ppm) : 1499.4476120257361
Photon noise performance (%) : 95.44547467386705
White noise (ppm) : 1570.9991669581843
Red noise (ppm) : 0.0
Transit depth measured error (ppm) : 70.04274835488246
/tmp/ipykernel_1956/3968974178.py:138: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:142: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.77921338 -0.73484018 0.59159019 -0.19763154
3param 1.86887525 -2.68691313 1.18957342
Quad 0.02139147 0.31022264
Linear 0.28448893
c1 = 0.7792133777907014
c2 = -0.7348401843860144
c3 = 0.5915901917273957
c4 = -0.19763153511838683
Fitting Bin 8 Wavelength = 3.4045925866666678 Range= [ 3.3099063 : 3.4860291 ]
rprs: 0.08227189100304827 current chi^2= 9103.126452750907 scatter 0.0014868598464022912
Re-Fitting Bin 8 Wavelength = 3.4045925866666678 Range= [ 3.3099063 : 3.4860291 ]
redchi 1.112443657918967
chi2 9103.126452750907
nfree 8183
bic 945.0253314503971
aic 881.9271113248834
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08227189 +/- 2.1027e-05 (0.03%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 501292.576 +/- 28.8379224 (0.01%) (init = 501326.9)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: -0.03070645 +/- 0.00892025 (29.05%) (init = -0.03123646)
xsh: 7.1767e-05 +/- 7.2558e-05 (101.10%) (init = -4.314611e-05)
ysh: -7.0107e-04 +/- 6.1147e-05 (8.72%) (init = -0.0007549212)
x2sh: -2.8376e-04 +/- 1.4177e-04 (49.96%) (init = -0.0003371664)
y2sh: -1.8900e-04 +/- 1.3982e-04 (73.98%) (init = -0.000238624)
xysh: 4.8986e-04 +/- 2.7908e-04 (56.97%) (init = 0.0005530067)
comm: 4.4287e-04 +/- 5.7141e-05 (12.90%) (init = 0.0005872099)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9776
C(x2sh, xysh) = -0.9771
C(x2sh, y2sh) = +0.9328
C(rprs, f0) = +0.8501
C(rprs, Fslope) = -0.6688
C(Fslope, xsh) = -0.5224
C(f0, Fslope) = -0.4657
C(Fslope, comm) = -0.4321
C(xsh, ysh) = -0.3548
C(rprs, ysh) = +0.3432
C(Fslope, ysh) = -0.3082
C(f0, y2sh) = -0.2821
C(xsh, x2sh) = +0.2806
C(f0, x2sh) = -0.2547
C(rprs, xsh) = +0.2477
C(xsh, xysh) = -0.2451
C(rprs, comm) = +0.2288
C(f0, ysh) = +0.2269
C(f0, xysh) = +0.1828
C(xsh, y2sh) = +0.1689
C(f0, xsh) = +0.1547
C(Fslope, x2sh) = -0.1485
C(xsh, comm) = -0.1402
C(ysh, comm) = -0.1337
C(Fslope, xysh) = +0.1222
C(f0, comm) = +0.1168
C(y2sh, comm) = -0.1096
C(xysh, comm) = +0.1012
Residual standard deviation (ppm) : 1486.8598464022912
Photon noise (ppm) : 1412.3891198622528
Photon noise performance (%) : 94.99140912842371
White noise (ppm) : 1485.1862825953015
Red noise (ppm) : 70.66355613452959
Transit depth measured error (ppm) : 3.459847161666703
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.74674613 -0.70346448 0.56972787 -0.19098962
3param 1.79176273 -2.57217104 1.13841509
Quad 0.02245657 0.29733359
Linear 0.27462292
c1 = 0.7467461281818586
c2 = -0.703464475253341
c3 = 0.5697278747363782
c4 = -0.19098961530866335
Fitting Bin 9 Wavelength = 3.576083346666666 Range= [ 3.4860291 : 3.6536426 ]
rprs: 0.08252602288912203 current chi^2= 9276.73253099844 scatter 0.0013166537622119156
Re-Fitting Bin 9 Wavelength = 3.576083346666666 Range= [ 3.4860291 : 3.6536426 ]
redchi 1.1335205927417449
chi2 9276.73253099844
nfree 8184
bic 1090.7733083166083
aic 1034.686001538374
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08252602 +/- 2.8079e-04 (0.34%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 651858.992 +/- 28.9193771 (0.00%) (init = 652312.5)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -1.4617e-04 +/- 5.4854e-05 (37.53%) (init = -0.0001409193)
ysh: -2.3084e-04 +/- 5.1041e-05 (22.11%) (init = -0.0003514006)
x2sh: 2.9062e-04 +/- 1.2460e-04 (42.87%) (init = 0.0004020877)
y2sh: 2.9660e-04 +/- 1.2354e-04 (41.65%) (init = 0.0003425623)
xysh: -1.9799e-04 +/- 2.4527e-04 (123.88%) (init = -0.0003119663)
comm: 1.0111e-04 +/- 4.5457e-05 (44.96%) (init = 0.0002516143)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9819
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8122
C(xsh, ysh) = -0.6332
C(xsh, comm) = -0.4869
C(f0, x2sh) = -0.3871
C(f0, y2sh) = -0.3306
C(ysh, comm) = -0.3013
C(f0, xysh) = +0.2759
C(xsh, x2sh) = +0.2441
C(xsh, xysh) = -0.2147
C(rprs, x2sh) = -0.1950
C(xysh, comm) = +0.1792
C(xsh, y2sh) = +0.1772
C(x2sh, comm) = -0.1718
C(rprs, xsh) = -0.1683
C(y2sh, comm) = -0.1482
C(rprs, y2sh) = -0.1417
C(rprs, ysh) = +0.1408
C(f0, xsh) = -0.1225
C(rprs, xysh) = +0.1022
Residual standard deviation (ppm) : 1316.6537622119156
Photon noise (ppm) : 1238.5774523478374
Photon noise performance (%) : 94.07009556309521
White noise (ppm) : 1306.3149424296028
Red noise (ppm) : 164.9979756990808
Transit depth measured error (ppm) : 46.34440978535159
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.72352161 -0.69353431 0.56713532 -0.19083063
3param 1.72408895 -2.47704753 1.09722831
Quad 0.02204262 0.28542380
Linear 0.26410838
c1 = 0.7235216095387155
c2 = -0.6935343054246529
c3 = 0.56713531990293
c4 = -0.19083063355448535
Fitting Bin 10 Wavelength = 3.739628686666667 Range= [ 3.6536426 : 3.8137721 ]
rprs: 0.08096454885995075 current chi^2= 8850.135154072304 scatter 0.001147068098288207
Re-Fitting Bin 10 Wavelength = 3.739628686666667 Range= [ 3.6536426 : 3.8137721 ]
redchi 1.0813948135474467
chi2 8850.135154072304
nfree 8184
bic 705.1213399520565
aic 649.0340331738222
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08096455 +/- 2.4849e-04 (0.31%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 819096.202 +/- 31.6234031 (0.00%) (init = 819352.4)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 1.5290e-04 +/- 4.7801e-05 (31.26%) (init = 0.0002021917)
ysh: -7.7256e-05 +/- 4.4471e-05 (57.56%) (init = -0.0001052952)
x2sh: 1.7909e-04 +/- 1.0855e-04 (60.61%) (init = 0.0001836095)
y2sh: 1.2922e-04 +/- 1.0762e-04 (83.29%) (init = 0.0001099149)
xysh: -1.6232e-04 +/- 2.1368e-04 (131.64%) (init = -0.0001007594)
comm: -3.7415e-04 +/- 3.9600e-05 (10.58%) (init = -0.0003567791)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8117
C(xsh, ysh) = -0.6333
C(xsh, comm) = -0.4870
C(f0, x2sh) = -0.3874
C(f0, y2sh) = -0.3306
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2762
C(xsh, x2sh) = +0.2445
C(xsh, xysh) = -0.2149
C(rprs, x2sh) = -0.1952
C(xysh, comm) = +0.1794
C(xsh, y2sh) = +0.1774
C(x2sh, comm) = -0.1722
C(rprs, xsh) = -0.1700
C(y2sh, comm) = -0.1485
C(rprs, ysh) = +0.1418
C(rprs, y2sh) = -0.1416
C(f0, xsh) = -0.1239
C(rprs, xysh) = +0.1024
Residual standard deviation (ppm) : 1147.068098288207
Photon noise (ppm) : 1104.9243487157835
Photon noise performance (%) : 96.32595923159964
White noise (ppm) : 1143.761751956722
Red noise (ppm) : 87.20013698637507
Transit depth measured error (ppm) : 40.238203628442655
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.70158183 -0.68497822 0.56261757 -0.18898808
3param 1.65933401 -2.38925471 1.06001228
Quad 0.02095753 0.27363515
Linear 0.25302542
c1 = 0.7015818333762096
c2 = -0.6849782191633046
c3 = 0.5626175733208771
c4 = -0.1889880833590714
Fitting Bin 11 Wavelength = 3.896151973333334 Range= [ 3.8137721 : 3.9672586 ]
rprs: 0.08035896954195282 current chi^2= 9206.088737155027 scatter 0.0012179575143038588
Re-Fitting Bin 11 Wavelength = 3.896151973333334 Range= [ 3.8137721 : 3.9672586 ]
redchi 1.1248886531225595
chi2 9206.088737155027
nfree 8184
bic 1028.151182735288
aic 972.0638759570537
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08035897 +/- 2.6526e-04 (0.33%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 755663.729 +/- 30.9654606 (0.00%) (init = 755688.6)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 3.6211e-04 +/- 5.0760e-05 (14.02%) (init = 0.0003654635)
ysh: 1.8443e-04 +/- 4.7221e-05 (25.60%) (init = 0.0001264471)
x2sh: -8.4928e-05 +/- 1.1525e-04 (135.70%) (init = -6.479008e-05)
y2sh: -1.0979e-04 +/- 1.1426e-04 (104.07%) (init = -0.0001160302)
xysh: 1.8924e-04 +/- 2.2687e-04 (119.88%) (init = 0.0001877641)
comm: -7.9893e-04 +/- 4.2044e-05 (5.26%) (init = -0.0007396911)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8116
C(xsh, ysh) = -0.6333
C(xsh, comm) = -0.4870
C(f0, x2sh) = -0.3875
C(f0, y2sh) = -0.3307
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2764
C(xsh, x2sh) = +0.2447
C(xsh, xysh) = -0.2150
C(rprs, x2sh) = -0.1953
C(xysh, comm) = +0.1796
C(xsh, y2sh) = +0.1775
C(x2sh, comm) = -0.1724
C(rprs, xsh) = -0.1709
C(y2sh, comm) = -0.1487
C(rprs, ysh) = +0.1423
C(rprs, y2sh) = -0.1415
C(f0, xsh) = -0.1246
C(rprs, xysh) = +0.1026
Residual standard deviation (ppm) : 1217.9575143038587
Photon noise (ppm) : 1150.365136647971
Photon noise performance (%) : 94.4503501261683
White noise (ppm) : 1217.9575143038587
Red noise (ppm) : 0.0
Transit depth measured error (ppm) : 42.632166958260974
/tmp/ipykernel_1956/3968974178.py:138: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:142: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.70601642 -0.70030861 0.58051914 -0.19676969
3param 1.65882163 -2.39001144 1.06012540
Quad 0.01948102 0.27394442
Linear 0.25181120
c1 = 0.7060164155107614
c2 = -0.700308605577277
c3 = 0.5805191372829517
c4 = -0.1967696897622868
Fitting Bin 12 Wavelength = 4.046415566666666 Range= [ 3.9672586 : 4.1148019 ]
rprs: 0.08042004711851963 current chi^2= 9426.689204343304 scatter 0.001324849605356413
Re-Fitting Bin 12 Wavelength = 4.046415566666666 Range= [ 3.9672586 : 4.1148019 ]
redchi 1.1518437444212248
chi2 9426.689204343304
nfree 8184
bic 1222.1365859827429
aic 1166.0492792045086
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08042005 +/- 2.8830e-04 (0.36%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 653878.858 +/- 29.1478519 (0.00%) (init = 653852)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 4.1610e-04 +/- 5.5217e-05 (13.27%) (init = 0.0003917553)
ysh: 2.5082e-04 +/- 5.1367e-05 (20.48%) (init = 0.0002782358)
x2sh: 9.2727e-07 +/- 1.2537e-04 (13519.85%) (init = -5.076081e-05)
y2sh: -5.9623e-05 +/- 1.2429e-04 (208.47%) (init = -8.608243e-05)
xysh: 5.8585e-06 +/- 2.4679e-04 (4212.40%) (init = 9.359886e-05)
comm: -8.7773e-04 +/- 4.5735e-05 (5.21%) (init = -0.0008647874)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8116
C(xsh, ysh) = -0.6333
C(xsh, comm) = -0.4870
C(f0, x2sh) = -0.3876
C(f0, y2sh) = -0.3307
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2764
C(xsh, x2sh) = +0.2446
C(xsh, xysh) = -0.2149
C(rprs, x2sh) = -0.1953
C(xysh, comm) = +0.1795
C(xsh, y2sh) = +0.1775
C(x2sh, comm) = -0.1724
C(rprs, xsh) = -0.1709
C(y2sh, comm) = -0.1487
C(rprs, ysh) = +0.1423
C(rprs, y2sh) = -0.1416
C(f0, xsh) = -0.1246
C(rprs, xysh) = +0.1026
Residual standard deviation (ppm) : 1324.849605356413
Photon noise (ppm) : 1236.6629569412296
Photon noise performance (%) : 93.34364836139576
White noise (ppm) : 1324.849605356413
Red noise (ppm) : 0.0
Transit depth measured error (ppm) : 46.36947542157553
/tmp/ipykernel_1956/3968974178.py:138: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:142: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.70935323 -0.72091003 0.59985926 -0.20291388
3param 1.64937007 -2.38471082 1.05992162
Quad 0.01796001 0.27118947
Linear 0.24795374
c1 = 0.7093532347748518
c2 = -0.7209100290636149
c3 = 0.5998592638096363
c4 = -0.2029138849439886
Fitting Bin 13 Wavelength = 4.191056326666667 Range= [ 4.1148019 : 4.2569868 ]
rprs: 0.08076799980124444 current chi^2= 10166.76684680261 scatter 0.001923789469298889
Re-Fitting Bin 13 Wavelength = 4.191056326666667 Range= [ 4.1148019 : 4.2569868 ]
redchi 1.2422735638810618
chi2 10166.76684680261
nfree 8184
bic 1841.2822564792052
aic 1785.194949700971
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08076800 +/- 4.1673e-04 (0.52%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 334548.499 +/- 21.6569457 (0.01%) (init = 334610.8)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 0.00255080 +/- 8.0179e-05 (3.14%) (init = 0.002471999)
ysh: -1.7229e-04 +/- 7.4585e-05 (43.29%) (init = -0.0001798177)
x2sh: 2.3572e-04 +/- 1.8204e-04 (77.22%) (init = 0.000255704)
y2sh: 1.2318e-04 +/- 1.8048e-04 (146.52%) (init = 0.0001410066)
xysh: -3.1019e-04 +/- 3.5834e-04 (115.52%) (init = -0.0003726413)
comm: -0.00237086 +/- 6.6405e-05 (2.80%) (init = -0.002301265)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8117
C(xsh, ysh) = -0.6332
C(xsh, comm) = -0.4870
C(f0, x2sh) = -0.3876
C(f0, y2sh) = -0.3309
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2765
C(xsh, x2sh) = +0.2455
C(xsh, xysh) = -0.2157
C(rprs, x2sh) = -0.1954
C(xysh, comm) = +0.1799
C(xsh, y2sh) = +0.1783
C(x2sh, comm) = -0.1729
C(rprs, xsh) = -0.1715
C(y2sh, comm) = -0.1491
C(rprs, ysh) = +0.1425
C(rprs, y2sh) = -0.1419
C(f0, xsh) = -0.1256
C(rprs, xysh) = +0.1028
Residual standard deviation (ppm) : 1923.789469298889
Photon noise (ppm) : 1728.9023194239408
Photon noise performance (%) : 89.86962175513034
White noise (ppm) : 1923.789469298889
Red noise (ppm) : 0.0
Transit depth measured error (ppm) : 67.31755156120067
/tmp/ipykernel_1956/3968974178.py:138: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:142: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.66323587 -0.66364687 0.56071310 -0.19136859
3param 1.55253360 -2.22982036 0.98936587
Quad 0.02366143 0.25528761
Linear 0.24016891
c1 = 0.6632358693959266
c2 = -0.6636468698454744
c3 = 0.5607130986026022
c4 = -0.1913685852532563
Fitting Bin 14 Wavelength = 4.33061144 Range= [ 4.2569868 : 4.3943119 ]
rprs: 0.08114760991530912 current chi^2= 10258.192606422173 scatter 0.0016628191173252076
Re-Fitting Bin 14 Wavelength = 4.33061144 Range= [ 4.2569868 : 4.3943119 ]
redchi 1.253444844381986
chi2 10258.192606422173
nfree 8184
bic 1914.6204510224768
aic 1858.5331442442425
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08114761 +/- 3.5865e-04 (0.44%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 451670.955 +/- 25.2930094 (0.01%) (init = 451633.6)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: -5.6351e-04 +/- 6.9301e-05 (12.30%) (init = -0.0005649959)
ysh: 5.0939e-05 +/- 6.4477e-05 (126.57%) (init = -4.022727e-05)
x2sh: 2.6615e-05 +/- 1.5736e-04 (591.25%) (init = -6.50098e-06)
y2sh: 2.7290e-05 +/- 1.5601e-04 (571.68%) (init = -4.604367e-05)
xysh: -1.3847e-04 +/- 3.0976e-04 (223.71%) (init = -5.075544e-05)
comm: 2.9164e-04 +/- 5.7406e-05 (19.68%) (init = 0.0003705078)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8119
C(xsh, ysh) = -0.6334
C(xsh, comm) = -0.4869
C(f0, x2sh) = -0.3876
C(f0, y2sh) = -0.3309
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2764
C(xsh, x2sh) = +0.2443
C(xsh, xysh) = -0.2147
C(rprs, x2sh) = -0.1954
C(xysh, comm) = +0.1791
C(xsh, y2sh) = +0.1772
C(x2sh, comm) = -0.1718
C(rprs, xsh) = -0.1703
C(y2sh, comm) = -0.1482
C(rprs, ysh) = +0.1424
C(rprs, y2sh) = -0.1420
C(f0, xsh) = -0.1240
C(rprs, xysh) = +0.1028
Residual standard deviation (ppm) : 1662.8191173252076
Photon noise (ppm) : 1487.9519878305853
Photon noise performance (%) : 89.48369502896313
White noise (ppm) : 1661.4245919556586
Red noise (ppm) : 68.21913628208945
Transit depth measured error (ppm) : 58.207801105886354
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.60983174 -0.61629105 0.53858322 -0.18657597
3param 1.42144129 -2.02725487 0.89908507
Quad 0.03065179 0.23249928
Linear 0.22783267
c1 = 0.6098317351531414
c2 = -0.6162910548658452
c3 = 0.5385832222369871
c4 = -0.1865759656282499
Fitting Bin 15 Wavelength = 4.465538639999999 Range= [ 4.3943119 : 4.5272022 ]
rprs: 0.08073844912626177 current chi^2= 10929.456415288656 scatter 0.0018686202314533622
Re-Fitting Bin 15 Wavelength = 4.465538639999999 Range= [ 4.3943119 : 4.5272022 ]
redchi 1.3354663264037947
chi2 10929.456415288656
nfree 8184
bic 2433.86957368386
aic 2377.7822669056254
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08073845 +/- 4.0441e-04 (0.50%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 381159.922 +/- 23.9803751 (0.01%) (init = 381273.9)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 8.1010e-04 +/- 7.7894e-05 (9.62%) (init = 0.0008546914)
ysh: -0.00148161 +/- 7.2466e-05 (4.89%) (init = -0.001520144)
x2sh: 4.3154e-04 +/- 1.7687e-04 (40.99%) (init = 0.0004347958)
y2sh: 4.8933e-04 +/- 1.7536e-04 (35.84%) (init = 0.0004636312)
xysh: -8.5004e-04 +/- 3.4817e-04 (40.96%) (init = -0.0008122209)
comm: 4.8510e-04 +/- 6.4511e-05 (13.30%) (init = 0.0004869507)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9378
C(rprs, f0) = +0.8119
C(xsh, ysh) = -0.6335
C(xsh, comm) = -0.4867
C(f0, x2sh) = -0.3877
C(f0, y2sh) = -0.3311
C(ysh, comm) = -0.3011
C(f0, xysh) = +0.2767
C(xsh, x2sh) = +0.2452
C(xsh, xysh) = -0.2155
C(rprs, x2sh) = -0.1957
C(xysh, comm) = +0.1791
C(xsh, y2sh) = +0.1781
C(x2sh, comm) = -0.1718
C(rprs, xsh) = -0.1715
C(y2sh, comm) = -0.1482
C(rprs, ysh) = +0.1437
C(rprs, y2sh) = -0.1422
C(f0, xsh) = -0.1253
C(rprs, xysh) = +0.1031
Residual standard deviation (ppm) : 1868.6202314533623
Photon noise (ppm) : 1619.7440194789522
Photon noise performance (%) : 86.6812845229209
White noise (ppm) : 1868.6202314533623
Red noise (ppm) : 0.0
Transit depth measured error (ppm) : 65.30327824768023
/tmp/ipykernel_1956/3968974178.py:138: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sig0**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:142: RuntimeWarning: invalid value encountered in sqrt
sigrednoise = np.sqrt(sig_binned**2-sigwhite**2/binmeasure)
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.60997947 -0.64472844 0.57190253 -0.19852233
3param 1.39349756 -1.99455717 0.88740172
Quad 0.03077627 0.22593125
Linear 0.22238684
c1 = 0.6099794722510968
c2 = -0.6447284371809401
c3 = 0.5719025262011778
c4 = -0.19852232970661113
Fitting Bin 16 Wavelength = 4.59623168 Range= [ 4.5272022 : 4.6560258 ]
rprs: 0.08025350816232052 current chi^2= 11816.076602419895 scatter 0.002240984638087443
Re-Fitting Bin 16 Wavelength = 4.59623168 Range= [ 4.5272022 : 4.6560258 ]
redchi 1.4438021263953928
chi2 11816.076602419895
nfree 8184
bic 3072.8411538775026
aic 3016.753847099268
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.08025351 +/- 4.8728e-04 (0.61%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 286515.192 +/- 21.6081512 (0.01%) (init = 286539.3)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 0.00105521 +/- 9.3415e-05 (8.85%) (init = 0.0009579191)
ysh: -1.9211e-04 +/- 8.6901e-05 (45.23%) (init = -0.0002364105)
x2sh: 2.6687e-04 +/- 2.1207e-04 (79.47%) (init = 0.000171836)
y2sh: 3.2514e-04 +/- 2.1025e-04 (64.67%) (init = 0.000207163)
xysh: -6.0995e-04 +/- 4.1745e-04 (68.44%) (init = -0.0003703153)
comm: -9.2834e-04 +/- 7.7357e-05 (8.33%) (init = -0.0007643483)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8117
C(xsh, ysh) = -0.6335
C(xsh, comm) = -0.4869
C(f0, x2sh) = -0.3879
C(f0, y2sh) = -0.3311
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2768
C(xsh, x2sh) = +0.2451
C(xsh, xysh) = -0.2153
C(rprs, x2sh) = -0.1957
C(xysh, comm) = +0.1795
C(xsh, y2sh) = +0.1779
C(x2sh, comm) = -0.1724
C(rprs, xsh) = -0.1721
C(y2sh, comm) = -0.1486
C(rprs, ysh) = +0.1435
C(rprs, y2sh) = -0.1421
C(f0, xsh) = -0.1258
C(rprs, xysh) = +0.1031
Residual standard deviation (ppm) : 2240.984638087443
Photon noise (ppm) : 1868.212066695138
Photon noise performance (%) : 83.36567930646568
White noise (ppm) : 2239.910337649141
Red noise (ppm) : 69.5171306214575
Transit depth measured error (ppm) : 78.21205389847466
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
You are using the 3D limb darkening models.
Current Directories Entered:
./notebookrun2/
./notebookrun2/3DGrid
Filename: mmu_t45g45m00v05.flx
Closest values to your inputs:
Teff : 4500
M_H : 0.0
log(g): 4.5
Limb darkening parameters:
4param 0.60350262 -0.62606713 0.54814049 -0.18913089
3param 1.39051667 -1.99106813 0.88526265
Quad 0.02900564 0.22612279
Linear 0.22077866
c1 = 0.6035026150544636
c2 = -0.6260671293320387
c3 = 0.5481404929821063
c4 = -0.18913089064980246
Fitting Bin 17 Wavelength = 4.7230313 Range= [ 4.6560258 : 4.7811009 ]
rprs: 0.07993813095540425 current chi^2= 16280.793120699353 scatter 0.003335098304838922
Re-Fitting Bin 17 Wavelength = 4.7230313 Range= [ 4.6560258 : 4.7811009 ]
redchi 1.9893442229593539
chi2 16280.793120699353
nfree 8184
bic 5698.5823530513335
aic 5642.495046273099
L-M FIT Variable
[[Variables]]
cosinc: 0.05481076 (fixed)
rho_star: 8.320569 (fixed)
a_Rs: 14.54 (fixed)
rprs: 0.07993813 +/- 7.2749e-04 (0.91%) (init = 0.0804)
t0: 2454865 (fixed)
f0: 178202.655 +/- 19.9965859 (0.01%) (init = 178209.5)
ecc: 0 (fixed)
omega: 0 (fixed)
Fslope: 0 (fixed)
xsh: 0.00131928 +/- 1.3905e-04 (10.54%) (init = 0.001501862)
ysh: 5.1327e-05 +/- 1.2935e-04 (252.00%) (init = -1.888577e-05)
x2sh: 3.9976e-04 +/- 3.1563e-04 (78.95%) (init = 0.0001132571)
y2sh: 5.5004e-04 +/- 3.1294e-04 (56.89%) (init = 0.0002763527)
xysh: -0.00101505 +/- 6.2132e-04 (61.21%) (init = -0.000378254)
comm: -0.00132254 +/- 1.1513e-04 (8.71%) (init = -0.001357975)
[[Correlations]] (unreported correlations are < 0.100)
C(y2sh, xysh) = -0.9820
C(x2sh, xysh) = -0.9756
C(x2sh, y2sh) = +0.9377
C(rprs, f0) = +0.8116
C(xsh, ysh) = -0.6335
C(xsh, comm) = -0.4869
C(f0, x2sh) = -0.3879
C(f0, y2sh) = -0.3311
C(ysh, comm) = -0.3010
C(f0, xysh) = +0.2769
C(xsh, x2sh) = +0.2453
C(xsh, xysh) = -0.2154
C(rprs, x2sh) = -0.1957
C(xysh, comm) = +0.1796
C(xsh, y2sh) = +0.1780
C(x2sh, comm) = -0.1725
C(rprs, xsh) = -0.1725
C(y2sh, comm) = -0.1488
C(rprs, ysh) = +0.1437
C(rprs, y2sh) = -0.1420
C(f0, xsh) = -0.1262
C(rprs, xysh) = +0.1032
Residual standard deviation (ppm) : 3335.098304838922
Photon noise (ppm) : 2368.879199856751
Photon noise performance (%) : 71.02876687082129
White noise (ppm) : 3332.548577726672
Red noise (ppm) : 130.64106910830395
Transit depth measured error (ppm) : 116.3089626329386
/tmp/ipykernel_1956/3968974178.py:175: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax1.scatter(x, y/p['f0'].value, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
/tmp/ipykernel_1956/3968974178.py:190: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax2.scatter(x, residppm, s=msize*0.75, linewidth=1, alpha=0.5, marker='+', edgecolors='blue', zorder=0) # overplot Transit model at data
/tmp/ipykernel_1956/3968974178.py:216: UserWarning: You passed a edgecolor/edgecolors ('blue') for an unfilled marker ('+'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
plt.scatter(x, light_curve+resid, s=msize*0.75, linewidth=1, zorder=0, alpha=0.5, marker='+', edgecolors='blue')
** Can Now View Output PDFs in ./notebookrun2/
Plot Measured Exoplanet Transmission Spectrum vs Injected#
# --------------------------------------------------------------------------
# Load Injected Transmission spectra to compare with recovered value
# Download Injected Spectra
fn_tm = download_file(r'https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/transit_spectroscopy_notebook/trans-iso_GJ-0436_0669.0_+0.0_0.56_0001_0.00_model.NIRSpec_PRISM.txt')
destld = shutil.move(fn_tm, save_directory+'trans-iso_GJ-0436_0669.0_+0.0_0.56_0001_0.00_model.NIRSpec_PRISM.txt')
f = open(save_directory+'trans-iso_GJ-0436_0669.0_+0.0_0.56_0001_0.00_model.NIRSpec_PRISM.txt', 'r')
data = np.genfromtxt(f, delimiter=' ')
model_ws = data[:, 0]
model_spec = data[:, 1]
# Read fit transit depths
data = ascii.read(save_directory+'JWST_NIRSpec_Prism_fit_transmission_spectra.csv', format='csv')
wsd = data['Wavelength Center (um)']
werr = data['Wavelength half-width (um)']
rprs = data['Rp/Rs']
rerr = data['Rp/Rs 1-sigma error']
beta = data['Beta Rednoise Inflation factor']
# plot
fig, axs = plt.subplots()
plt.plot(model_ws, model_spec**2*1E6, linewidth=2, zorder=0, color='blue', label='Injected Spectra') # overplot Transit model at data
plt.errorbar(wsd, rprs**2*1E6, xerr=werr, yerr=2*rerr*rprs*1E6*beta, fmt='o', zorder=5, alpha=0.4, color='orange', label=r'Recovered Spectra with $\sigma_r$')
plt.errorbar(wsd, rprs**2*1E6, xerr=werr, yerr=2*rerr*rprs*1E6, fmt='o', zorder=10, color='orange', label='Recovered Spectra')
plt.xlabel(r'Wavelength ($\mu$m)')
plt.ylabel('Transit Depth ($R_p/R_s$)$^2$ (ppm)')
axs.yaxis.set_major_locator(ticker.MultipleLocator(200))
axs.yaxis.set_minor_locator(ticker.MultipleLocator(100))
axs.xaxis.set_major_locator(ticker.MultipleLocator(0.5))
axs.xaxis.set_minor_locator(ticker.MultipleLocator(0.1))
axs.text(3.3, 6850, 'CH$_4$')
axs.text(4.25, 6700, 'CO')
axs.text(2.3, 6750, 'CH$_4$')
axs.text(2.75, 6550, 'H$_2$O')
plt.ylim(5700, 7000)
plt.xlim(0.9, 5.25)
plt.legend(loc='lower right')
plt.show()
plt.clf()
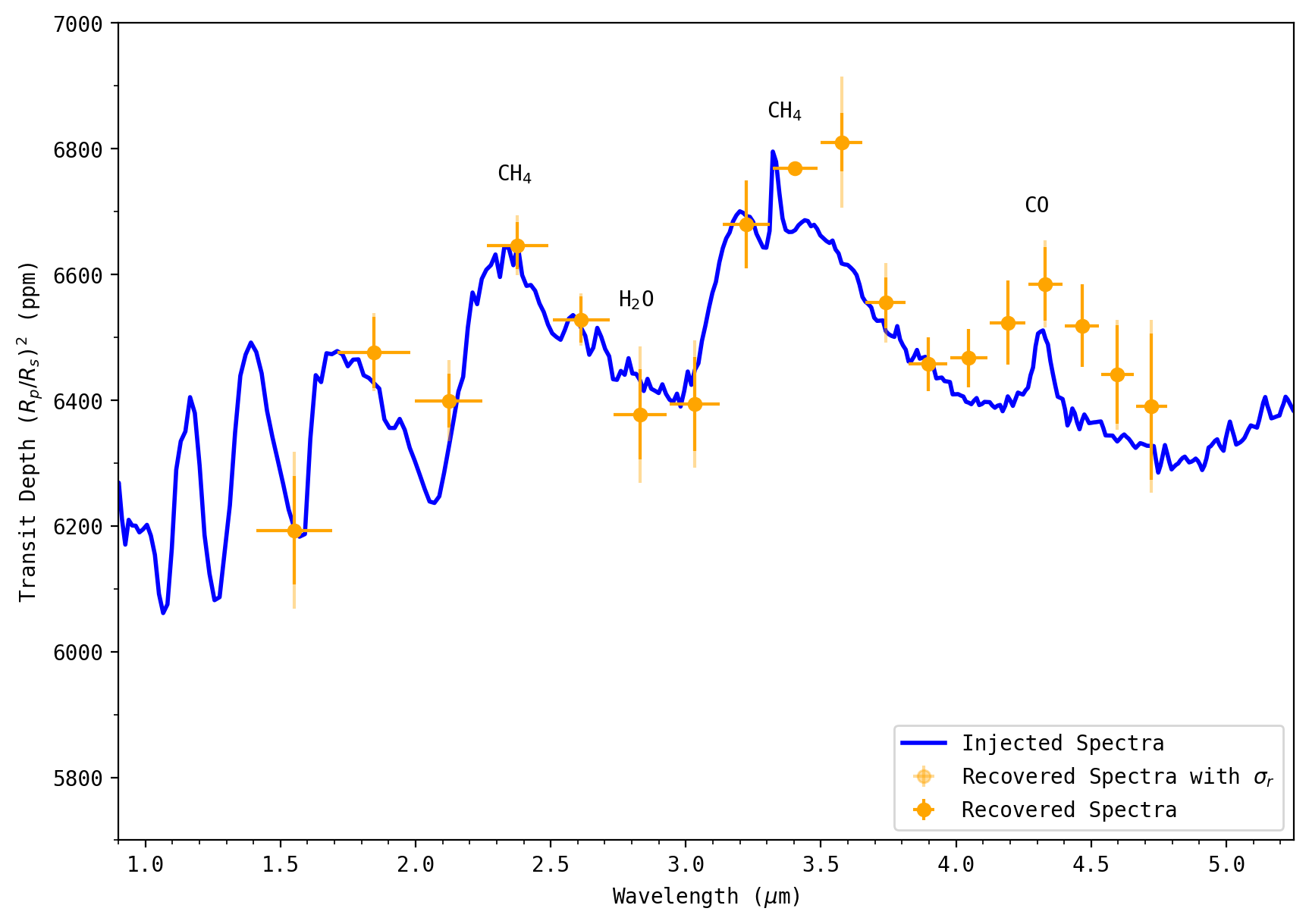
<Figure size 2000x1400 with 0 Axes>
By and large, the injected transit depths are well recovered across the spectra, with features such as H\(_2\)O and CH\(_4\) easily detected. There is a bit of an offset in data-points long-ward of 3.5 \(\mu\)m that could perhaps be due to changes in CO\(_2\) or H\(_2\)O absorption features from ice built up on the cryogenic window during the CV3 test. These wavelengths show some increases in time correlated noise (\(\sigma_r\)), which has been measured here, and the errors in the plot also show the transit depths with this error included.
The precisions from the ground-based test are very encouraging, with the best measured bin (which occurs in a clean part of the spectrum with high count rates) achieving near-photon limited transit depths measured to about 30 ppm in only 2 hours of data, and with minimal time correlated noise (\(\sigma_r\)).
For more robust error estimates, in practice the least-squares minimization performed here would be replaced by an MCMC routine. In addition, with actual transit data, the transit fit parameters (e.g. \(i\), \(a/R_{star}\), T\(_0\)) would also have to be first fit as well, as they can/will differ from literature estimates in high precision transit light curves as JWST will provide.
