Extended Aperture Photometry#
Use case: Measure galaxy photometry in a field. Related to JDox Science Use Case #22.
Data: WST simulated NIRCam images from JADES JWST GTO extragalactic blank field.
(Williams et al. 2018) https://ui.adsabs.harvard.edu/abs/2018ApJS..236…33W.
Tools: photutils, matplotlib, scipy, scikit.
Cross-intrument: potentially MIRI.
Documentation: This notebook is part of a STScI’s larger post-pipeline Data Analysis Tools Ecosystem.
Introduction#
This notebook uses photutils to detect objects/galaxies in NIRCam deep imaging. Detections are first made in a F200W image, then isophotal photometry is obtained in all 9 filters (F090W, F115W, F150W, F200W, F277W, F335M, F356W, F410M, F444W). The catalog is loaded back in and some simple analysis is performed on the full catalog and on an individual galaxy.
The notebook analyzes only the central 1000 x 1000 pixels (30” x 30”) of the full JADES simulation. These cutouts have been staged at STScI with permission from the authors (Williams et al.).
NOTE: The photometry is aperture matched, but no PSF corrections are made. For more accurate color measurements, PSF corrections should be implemented, given the large range of wavelengths (and thus PSF FWHM) spanning a factor of >4.
NOTE: The simulated JADES images have different units (e-/s) than JWST pipeline products (MJy/sr).
NOTE: An exposure map is missing but required to calculate flux uncertainties.
To Do#
PSF corrections
Check accuracy of photometry against simulated JADES catalog
Exposure map required for input to error calculation
ABmag units cannot be written to ecsv file (astropy update coming soon)
plot with text labels looks horrible (I wish cursor hover would show id number instead)
Fix plot secondary axis: mag vs. flux
requirements.txt file – but I don’t know what versions are “required”
rest of Robel’s comments: https://github.com/spacetelescope/dat_pyinthesky/pull/82#pullrequestreview-355206337
Download WEBBPSF Data#
import os
import tarfile
import urllib.request
boxlink = 'https://stsci.box.com/shared/static/34o0keicz2iujyilg4uz617va46ks6u9.gz'
boxfile = './webbpsf-data/webbpsf-data-1.0.0.tar.gz'
webbpsf_folder = './webbpsf-data'
# Check whether the specified path exists or not
isExist = os.path.exists(webbpsf_folder)
if not isExist:
# Create a new directory because it does not exist
os.makedirs(webbpsf_folder)
urllib.request.urlretrieve(boxlink, boxfile)
gzf = tarfile.open(boxfile)
gzf.extractall(webbpsf_folder, filter='data') # add filter='data' to skip absolute (/) or relative (..) paths
Import packages#
import os
import numpy as np
from astropy.convolution import convolve
from astropy.io import fits
from astropy.stats import gaussian_fwhm_to_sigma
from astropy.table import QTable
import astropy.units as u
from astropy.visualization import make_lupton_rgb, SqrtStretch, ImageNormalize, simple_norm
import astropy.wcs as wcs
from photutils.background import Background2D, MedianBackground
from photutils.segmentation import (detect_sources, deblend_sources, SourceCatalog,
make_2dgaussian_kernel, SegmentationImage)
from photutils.utils import calc_total_error
Matplotlib setup for plotting#
There are two versions
notebook– gives interactive plots, but makes the overall notebook a bit harder to scrollinline– gives non-interactive plots for better overall scrolling
import matplotlib.pyplot as plt
import matplotlib as mpl
# Use this version for non-interactive plots (easier scrolling of the notebook)
%matplotlib inline
# Use this version if you want interactive plots
# %matplotlib notebook
# These gymnastics are needed to make the sizes of the figures
# be the same in both the inline and notebook versions
%config InlineBackend.print_figure_kwargs = {'bbox_inches': None}
mpl.rcParams['savefig.dpi'] = 80
mpl.rcParams['figure.dpi'] = 80
Create list of images to be loaded and analyzed#
baseurl = 'https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/'
filters = 'F090W F115W F150W F200W F277W F335M F356W F410M F444W'.split()
# Data images [e-/s]
imagefiles = {}
for filt in filters:
filename = f'jades_jwst_nircam_goods_s_crop_{filt}.fits'
imagefiles[filt] = os.path.join(baseurl, filename)
# Weight images (Inverse Variance Maps; IVM)
weightfiles = {}
for filt in filters:
filename = f'jades_jwst_nircam_goods_s_crop_{filt}_wht.fits'
weightfiles[filt] = os.path.join(baseurl, filename)
Load detection image: F200W#
filt = 'F200W'
infile = imagefiles[filt]
hdu = fits.open(infile)
data = hdu[0].data
imwcs = wcs.WCS(hdu[0].header, hdu)
weight = fits.open(weightfiles[filt])[0].data
Report image size and field of view#
ny, nx = data.shape
pixscale = wcs.utils.proj_plane_pixel_scales(imwcs)[0]
pixscale *= imwcs.wcs.cunit[0].to('arcsec')
outline = '%d x %d pixels' % (ny, nx)
outline += ' = %g" x %g"' % (ny * pixscale, nx * pixscale)
outline += ' (%.2f" / pixel)' % pixscale
print(outline)
1000 x 1000 pixels = 30" x 30" (0.03" / pixel)
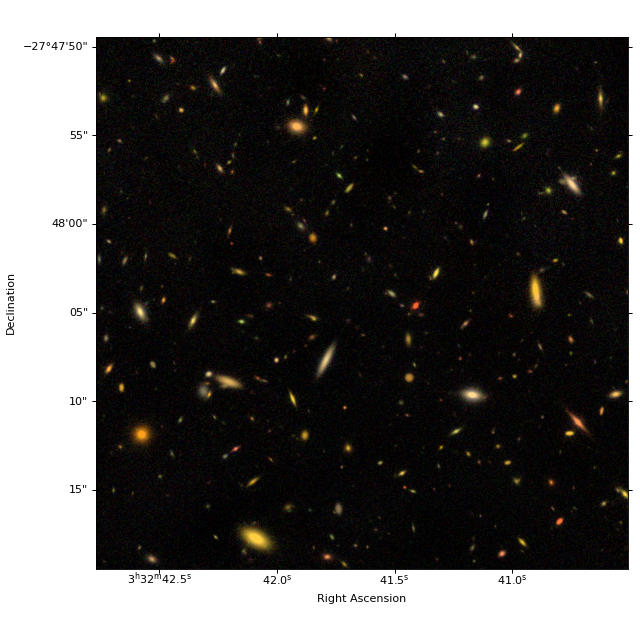
Create color image (optional)#
# 3 NIRCam short wavelength channel images
r = fits.open(imagefiles['F200W'])[0].data
g = fits.open(imagefiles['F150W'])[0].data
b = fits.open(imagefiles['F090W'])[0].data
rgb = make_lupton_rgb(r, g, b, Q=5, stretch=0.02) # , minimum=-0.001
fig = plt.figure(figsize=(8, 8))
ax = plt.subplot(projection=imwcs)
plt.imshow(rgb, origin='lower')
plt.xlabel('Right Ascension')
plt.ylabel('Declination')
fig.tight_layout()
plt.subplots_adjust(left=0.15)
Detect Sources and Deblend using photutils#
https://photutils.readthedocs.io/en/latest/segmentation.html
# For detection, requiring 5 connected pixels 2-sigma above background
# Measure background and set detection threshold
bkg_estimator = MedianBackground()
bkg = Background2D(data, (50, 50), filter_size=(3, 3), bkg_estimator=bkg_estimator)
threshold = bkg.background + (2. * bkg.background_rms)
# Before detection, smooth image with Gaussian FWHM = 3 pixels
smooth_kernel = make_2dgaussian_kernel(3.0, size=3)
convolved_data = convolve(data, smooth_kernel)
# Detect and deblend
segm_detect = detect_sources(convolved_data, threshold, npixels=5)
segm_deblend = deblend_sources(convolved_data, segm_detect, npixels=5, nlevels=32, contrast=0.001)
# Save segmentation map of detected objects
segm_hdu = fits.PrimaryHDU(segm_deblend.data.astype(np.uint32), header=imwcs.to_header())
segm_hdu.writeto('JADES_detections_segm.fits', overwrite=True)

Measure photometry (and more) in detection image#
https://photutils.readthedocs.io/en/latest/segmentation.html#centroids-photometry-and-morphological-properties
#error = bkg.background_rms
# Input weight should be exposure map. Fudging for now.
error = calc_total_error(data, bkg.background_rms, weight/500)
#cat = source_properties(data-bkg.background, segm_deblend, wcs=imwcs, background=bkg.background, error=error)
cat = SourceCatalog(data-bkg.background, segm_deblend, wcs=imwcs, background=bkg.background, error=error)
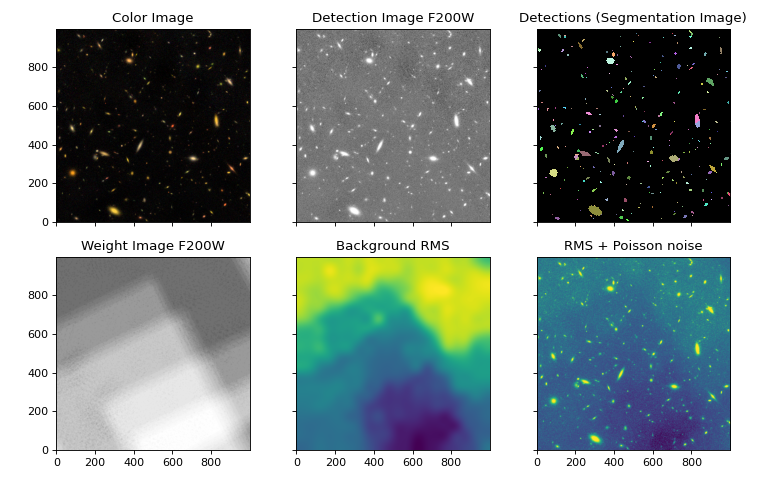
Show detections alongside images (optional)#
fig, ax = plt.subplots(2, 3, sharex=True, sharey=True, figsize=(9.5, 6))
# For RA,Dec axes instead of pixels, add: , subplot_kw={'projection': imwcs})
# Color image
ax[0, 0].imshow(rgb, origin='lower')
ax[0, 0].set_title('Color Image')
# Data
norm = simple_norm(data, 'sqrt', percent=99.)
ax[0, 1].imshow(data, origin='lower', cmap='Greys_r', norm=norm)
ax[0, 1].set_title('Detection Image F200W')
# Segmentation map
cmap = segm_deblend.make_cmap(seed=12345)
ax[0, 2].imshow(segm_deblend, origin='lower', cmap=cmap, interpolation='nearest')
ax[0, 2].set_title('Detections (Segmentation Image)')
# Weight
ax[1, 0].imshow(weight, origin='lower', cmap='Greys_r', vmin=0)
ax[1, 0].set_title('Weight Image F200W')
# RMS
ax[1, 1].imshow(bkg.background_rms, origin='lower', norm=None)
ax[1, 1].set_title('Background RMS')
# Total error including Poisson noise
norm = simple_norm(error, 'sqrt', percent=99.)
ax[1, 2].imshow(error, origin='lower', norm=norm)
ax[1, 2].set_title('RMS + Poisson noise')
fig.tight_layout()
View all measured quantities in detection image (optional)#
cat.to_table()
| label | xcentroid | ycentroid | sky_centroid | bbox_xmin | bbox_xmax | bbox_ymin | bbox_ymax | area | semimajor_sigma | semiminor_sigma | orientation | eccentricity | min_value | max_value | local_background | segment_flux | segment_fluxerr | kron_flux | kron_fluxerr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg,deg | pix2 | pix | pix | deg | |||||||||||||||
| int32 | float64 | float64 | SkyCoord | int64 | int64 | int64 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 466.24461350033647 | 8.7975642602371 | 53.17381009103326,-27.805320830365776 | 460 | 472 | 3 | 14 | 101.0 | 3.121883592723396 | 1.687718238373556 | -44.478947493786606 | 0.8412740063312978 | 0.0006697164628791134 | 0.03003565456943476 | 0.0 | 0.8863594626861904 | 0.04333105379347733 | 1.1523646398846878 | 0.05542864325311204 |
| 2 | 104.80681791619067 | 18.082129411035602 | 53.17721522973072,-27.805243193448323 | 95 | 116 | 10 | 27 | 271.0 | 4.4330760205526705 | 2.8904310038200527 | -26.54972210581966 | 0.7582062720085906 | 0.001169592682614256 | 0.05683289995260978 | 0.0 | 3.3850931796608226 | 0.10096273567434758 | 4.064307886560463 | 0.1178471432803697 |
| 3 | 15.04416396551755 | 13.21870964358083 | 53.178060898531776,-27.805283643045666 | 14 | 16 | 12 | 14 | 8.0 | 0.8501945455843772 | 0.6682678017840504 | -38.1612276642421 | 0.6182042301587309 | 0.0035728101756059504 | 0.006067336431199741 | 0.0 | 0.04072228096572398 | 0.011222825585360583 | 0.10086850698470742 | 0.02315230578296353 |
| 4 | 710.3242889701858 | 14.259777138086877 | 53.17151058505514,-27.805275444028442 | 708 | 712 | 12 | 17 | 25.0 | 1.2749753643104456 | 1.117371994707132 | -59.26178196559024 | 0.4816073152747641 | 0.0013530635966815864 | 0.015100956012602468 | 0.0 | 0.15496901383687856 | 0.018575211927226793 | 0.19011686546526946 | 0.023625589169720226 |
| 5 | 434.5294891713508 | 22.219091527997158 | 53.17410887322447,-27.8052089643638 | 424 | 447 | 14 | 30 | 268.0 | 3.9551276120227397 | 2.6287674709150584 | -8.140060370882344 | 0.7471566761152122 | -0.0003420958396561837 | 0.10656058422736421 | 0.0 | 3.6147863822998016 | 0.08773502088121837 | 4.278741209374845 | 0.10086436727140968 |
| 6 | 289.38802515014896 | 17.281765562153954 | 53.17547627056455,-27.805250009193262 | 287 | 291 | 15 | 20 | 17.0 | 1.2913044931147235 | 1.1978568863866517 | 68.12270923149859 | 0.37349256122504115 | 0.0019512862197305747 | 0.008797215945005498 | 0.0 | 0.08513804286457863 | 0.0166428570162664 | 0.34384198668200716 | 0.045989916939691816 |
| 7 | 640.4863091995317 | 17.773429154661983 | 53.17216853452775,-27.805246129692247 | 638 | 642 | 15 | 20 | 21.0 | 1.5124916692884618 | 0.8803106550282112 | -64.72678377811528 | 0.8131696183154983 | 0.0003576250881105088 | 0.009946935910809692 | 0.0 | 0.09708323832036175 | 0.014462494201596655 | 0.17695836262605436 | 0.026075310664432968 |
| 8 | 762.762291737285 | 28.289914155884627 | 53.171016553902064,-27.805158549648304 | 754 | 771 | 21 | 36 | 200.0 | 3.160315774736445 | 2.3536095208280066 | 37.520056617399476 | 0.6673561574146197 | -0.0001941792050655057 | 0.10556121967783572 | 0.0 | 3.8212752545169955 | 0.08847910158114891 | 3.9811392641181644 | 0.09150995786991047 |
| 9 | 801.0706722163886 | 32.58026118517374 | 53.17065564474934,-27.805122812753073 | 799 | 804 | 31 | 34 | 12.0 | 1.3349434908844906 | 0.6567170220834244 | -28.75519616047975 | 0.8706270208190223 | 0.0018856854900614819 | 0.009336017794193286 | 0.0 | 0.05935203501404956 | 0.012189355644992149 | 0.17003504172320322 | 0.027778386430149567 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 318 | 13.420742051379385 | 884.694395861645 | 53.17807529846922,-27.798021345154865 | 6 | 21 | 877 | 892 | 185.0 | 3.285450616131066 | 3.147908093244288 | -13.393044858704108 | 0.2863139002521662 | 0.0014361034419094758 | 0.03495382468946091 | 0.0 | 2.117442472888257 | 0.10785932211319861 | 3.411588966877565 | 0.15467709845637975 |
| 319 | 7.027176577444122 | 891.9727894960166 | 53.17813552144662,-27.797960686059632 | 5 | 11 | 889 | 895 | 29.0 | 1.603195468602836 | 1.3128724093575075 | -82.75208405786572 | 0.5739223050191917 | 0.0019018454919824644 | 0.01347560203904176 | 0.0 | 0.21412965179118829 | 0.035024475587970415 | 0.6893013683010651 | 0.07610234326451545 |
| 320 | 143.92969676102126 | 953.6973600108524 | 53.17684577662435,-27.797446433611444 | 139 | 149 | 950 | 957 | 59.0 | 2.6507581110195266 | 1.3964068465385502 | 25.19703083525805 | 0.849992199281631 | 0.0021617009219625062 | 0.021165230633174926 | 0.0 | 0.5681924075559929 | 0.05743803199966468 | 1.254760987505672 | 0.10492362090925443 |
| 321 | 142.746173256619 | 959.5389917866399 | 53.176856920479636,-27.797397752374 | 139 | 147 | 957 | 963 | 48.0 | 2.1427436610213637 | 1.2555061677914512 | -35.47427950196171 | 0.8103588434156908 | 0.00047583665796055224 | 0.030453184966657037 | 0.0 | 0.5637982114834339 | 0.0568718012708143 | 0.8893801784930943 | 0.08214060528713435 |
| 322 | 790.415367777511 | 955.3106960545537 | 53.17075559998907,-27.797433388936582 | 784 | 796 | 951 | 959 | 93.0 | 2.621691845359079 | 1.8772136125476881 | 5.157966396104574 | 0.6980681000951601 | 0.0005767950066928708 | 0.04023169644016939 | 0.0 | 1.115611206251331 | 0.07893942785469334 | 1.6521126492979301 | 0.10804543306519865 |
| 323 | 797.3397409211948 | 965.5478941851635 | 53.17069036467991,-27.797348081794517 | 792 | 802 | 958 | 974 | 121.0 | 3.0304070507432326 | 1.7633197784086392 | 79.51657917516623 | 0.8132778374227926 | 0.002333420221800177 | 0.08761686954182783 | 0.0 | 2.116916544882111 | 0.10637880974470741 | 2.8478971911033626 | 0.1315925489047444 |
| 324 | 790.7983558965504 | 979.5822296300561 | 53.17075198078638,-27.79723112632807 | 781 | 799 | 972 | 989 | 133.0 | 4.506451359665357 | 1.493045462994405 | -44.19824469199431 | 0.9435209394229406 | -0.001248849458808359 | 0.06943444193177313 | 0.0 | 2.002750597650743 | 0.10372107669936124 | 2.813577038594517 | 0.13630578304670982 |
| 325 | 307.7502590912509 | 989.993370703415 | 53.175302486237165,-27.797144093448775 | 305 | 311 | 987 | 993 | 37.0 | 1.707727984108737 | 1.1914329585464343 | -56.82602073196763 | 0.7164179156288306 | 0.0017972712412996086 | 0.030535860130823354 | 0.0 | 0.3875293679154765 | 0.046167097763049746 | 0.7369473920493916 | 0.07417814641467854 |
| 326 | 306.8506442261885 | 997.243644725512 | 53.1753109550711,-27.797083673855024 | 302 | 312 | 994 | 999 | 51.0 | 2.5021353669231527 | 1.4883885517566502 | 15.388098948641728 | 0.8038386961956926 | 0.002853197675265836 | 0.02047051119926145 | 0.0 | 0.5505454403242959 | 0.05460363335321445 | 0.7777606691520305 | 0.07156975624994025 |
Only keep some quantities#
columns = 'label xcentroid ycentroid sky_centroid area semimajor_sigma semiminor_sigma ellipticity orientation gini'.split()
tbl = cat.to_table(columns=columns)
tbl.rename_column('semimajor_sigma', 'a')
tbl.rename_column('semiminor_sigma', 'b')
tbl
| label | xcentroid | ycentroid | sky_centroid | area | a | b | ellipticity | orientation | gini |
|---|---|---|---|---|---|---|---|---|---|
| deg,deg | pix2 | pix | pix | deg | |||||
| int32 | float64 | float64 | SkyCoord | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 466.24461350033647 | 8.7975642602371 | 53.17381009103326,-27.805320830365776 | 101.0 | 3.121883592723396 | 1.687718238373556 | 0.4593910412587602 | -44.478947493786606 | 0.3750841422808698 |
| 2 | 104.80681791619067 | 18.082129411035602 | 53.17721522973072,-27.805243193448323 | 271.0 | 4.4330760205526705 | 2.8904310038200527 | 0.34798523859744157 | -26.54972210581966 | 0.4292034166747856 |
| 3 | 15.04416396551755 | 13.21870964358083 | 53.178060898531776,-27.805283643045666 | 8.0 | 0.8501945455843772 | 0.6682678017840504 | 0.21398248759086114 | -38.1612276642421 | 0.10521135944081007 |
| 4 | 710.3242889701858 | 14.259777138086877 | 53.17151058505514,-27.805275444028442 | 25.0 | 1.2749753643104456 | 1.117371994707132 | 0.12361287442487268 | -59.26178196559024 | 0.33737600483155716 |
| 5 | 434.5294891713508 | 22.219091527997158 | 53.17410887322447,-27.8052089643638 | 268.0 | 3.9551276120227397 | 2.6287674709150584 | 0.33535204706817323 | -8.140060370882344 | 0.5363680123723463 |
| 6 | 289.38802515014896 | 17.281765562153954 | 53.17547627056455,-27.805250009193262 | 17.0 | 1.2913044931147235 | 1.1978568863866517 | 0.07236682535090477 | 68.12270923149859 | 0.18041073594089005 |
| 7 | 640.4863091995317 | 17.773429154661983 | 53.17216853452775,-27.805246129692247 | 21.0 | 1.5124916692884618 | 0.8803106550282112 | 0.41797322067741016 | -64.72678377811528 | 0.257339933672325 |
| 8 | 762.762291737285 | 28.289914155884627 | 53.171016553902064,-27.805158549648304 | 200.0 | 3.160315774736445 | 2.3536095208280066 | 0.2552612812799423 | 37.520056617399476 | 0.5417393494467531 |
| 9 | 801.0706722163886 | 32.58026118517374 | 53.17065564474934,-27.805122812753073 | 12.0 | 1.3349434908844906 | 0.6567170220834244 | 0.5080563135685205 | -28.75519616047975 | 0.28793220448127643 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 318 | 13.420742051379385 | 884.694395861645 | 53.17807529846922,-27.798021345154865 | 185.0 | 3.285450616131066 | 3.147908093244288 | 0.041864127316802446 | -13.393044858704108 | 0.331692826709577 |
| 319 | 7.027176577444122 | 891.9727894960166 | 53.17813552144662,-27.797960686059632 | 29.0 | 1.603195468602836 | 1.3128724093575075 | 0.18109024440939903 | -82.75208405786572 | 0.254070174248954 |
| 320 | 143.92969676102126 | 953.6973600108524 | 53.17684577662435,-27.797446433611444 | 59.0 | 2.6507581110195266 | 1.3964068465385502 | 0.47320472557133164 | 25.19703083525805 | 0.2823870637944277 |
| 321 | 142.746173256619 | 959.5389917866399 | 53.176856920479636,-27.797397752374 | 48.0 | 2.1427436610213637 | 1.2555061677914512 | 0.41406609169806174 | -35.47427950196171 | 0.3421221334166607 |
| 322 | 790.415367777511 | 955.3106960545537 | 53.17075559998907,-27.797433388936582 | 93.0 | 2.621691845359079 | 1.8772136125476881 | 0.28396862664438105 | 5.157966396104574 | 0.37596577589838664 |
| 323 | 797.3397409211948 | 965.5478941851635 | 53.17069036467991,-27.797348081794517 | 121.0 | 3.0304070507432326 | 1.7633197784086392 | 0.4181244470190332 | 79.51657917516623 | 0.47716929153411286 |
| 324 | 790.7983558965504 | 979.5822296300561 | 53.17075198078638,-27.79723112632807 | 133.0 | 4.506451359665357 | 1.493045462994405 | 0.6686871012615841 | -44.19824469199431 | 0.42319913307021234 |
| 325 | 307.7502590912509 | 989.993370703415 | 53.175302486237165,-27.797144093448775 | 37.0 | 1.707727984108737 | 1.1914329585464343 | 0.3023286087519126 | -56.82602073196763 | 0.35854189153102795 |
| 326 | 306.8506442261885 | 997.243644725512 | 53.1753109550711,-27.797083673855024 | 51.0 | 2.5021353669231527 | 1.4883885517566502 | 0.4051526662256868 | 15.388098948641728 | 0.24216628412005226 |
Convert measured fluxes (data units) to magnitudes#
https://docs.astropy.org/en/stable/units/
https://docs.astropy.org/en/stable/units/equivalencies.html#photometric-zero-point-equivalency
https://docs.astropy.org/en/stable/units/logarithmic_units.html#logarithmic-units
# not detected: mag = 99; magerr = 1-sigma upper limit assuming zero flux
# not observed: mag = -99; magerr = 0
def fluxes2mags(flux, fluxerr):
nondet = flux < 0 # Non-detection if flux is negative
unobs = (fluxerr <= 0) + (fluxerr == np.inf) # Unobserved if flux uncertainty is negative or infinity
mag = flux.to(u.ABmag)
magupperlimit = fluxerr.to(u.ABmag) # 1-sigma upper limit if flux=0
mag = np.where(nondet, 99 * u.ABmag, mag)
mag = np.where(unobs, -99 * u.ABmag, mag)
magerr = 2.5 * np.log10(1 + fluxerr/flux)
magerr = magerr.value * u.ABmag
magerr = np.where(nondet, magupperlimit, magerr)
magerr = np.where(unobs, 0*u.ABmag, magerr)
return mag, magerr
# Includes features I couldn't find in astropy:
# mag = 99 / -99 for non-detections / unobserved
# flux uncertainties -> mag uncertainties
Multiband photometry using isophotal apertures defined in detection image#
(Similar to running SourceExtractor in double-image mode)
filters = 'F090W F115W F150W F200W F277W F335M F356W F410M F444W'.split()
for filt in filters:
infile = imagefiles[filt]
print(filt)
print(infile)
print(weightfiles[filt])
hdu = fits.open(infile)
data = hdu[0].data
zp = hdu[0].header['ABMAG'] * u.ABmag # zeropoint
weight = fits.open(weightfiles[filt])[0].data
# Measure background
bkg = Background2D(data, (50, 50), filter_size=(3, 3), bkg_estimator=bkg_estimator)
#error = bkg.background_rms
error = calc_total_error(data, bkg.background_rms, weight/500)
# Measure properties in each image of previously detected objects
filtcat = SourceCatalog(data-bkg.background, segm_deblend, wcs=imwcs, background=bkg.background, error=error)
# Convert measured fluxes to fluxes in nJy and to AB magnitudes
filttbl = filtcat.to_table()
tbl[filt+'_flux'] = flux = filttbl['segment_flux'] * zp.to(u.nJy)
tbl[filt+'_fluxerr'] = fluxerr = filttbl['segment_fluxerr'] * zp.to(u.nJy)
mag, magerr = fluxes2mags(flux, fluxerr)
#mag = mag * u.ABmag # incompatible with file writing
tbl[filt+'_mag'] = mag.value
tbl[filt+'_magerr'] = magerr.value
F090W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F090W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F090W_wht.fits
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/function/logarithmic.py:67: RuntimeWarning: invalid value encountered in log10
return dex.to(self._function_unit, np.log10(x))
F115W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F115W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F115W_wht.fits
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/function/logarithmic.py:67: RuntimeWarning: invalid value encountered in log10
return dex.to(self._function_unit, np.log10(x))
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/quantity.py:659: RuntimeWarning: invalid value encountered in log10
result = super().__array_ufunc__(function, method, *arrays, **kwargs)
F150W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F150W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F150W_wht.fits
F200W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F200W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F200W_wht.fits
F277W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F277W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F277W_wht.fits
F335M
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F335M.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F335M_wht.fits
F356W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F356W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F356W_wht.fits
F410M
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F410M.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F410M_wht.fits
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/function/logarithmic.py:67: RuntimeWarning: invalid value encountered in log10
return dex.to(self._function_unit, np.log10(x))
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/quantity.py:659: RuntimeWarning: invalid value encountered in log10
result = super().__array_ufunc__(function, method, *arrays, **kwargs)
F444W
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F444W.fits
https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/nircam_photometry/jades_jwst_nircam_goods_s_crop_F444W_wht.fits
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/function/logarithmic.py:67: RuntimeWarning: invalid value encountered in log10
return dex.to(self._function_unit, np.log10(x))
/opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/astropy/units/quantity.py:659: RuntimeWarning: invalid value encountered in log10
result = super().__array_ufunc__(function, method, *arrays, **kwargs)
View complete results (optional)#
tbl
| label | xcentroid | ycentroid | sky_centroid | area | a | b | ellipticity | orientation | gini | F090W_flux | F090W_fluxerr | F090W_mag | F090W_magerr | F115W_flux | F115W_fluxerr | F115W_mag | F115W_magerr | F150W_flux | F150W_fluxerr | F150W_mag | F150W_magerr | F200W_flux | F200W_fluxerr | F200W_mag | F200W_magerr | F277W_flux | F277W_fluxerr | F277W_mag | F277W_magerr | F335M_flux | F335M_fluxerr | F335M_mag | F335M_magerr | F356W_flux | F356W_fluxerr | F356W_mag | F356W_magerr | F410M_flux | F410M_fluxerr | F410M_mag | F410M_magerr | F444W_flux | F444W_fluxerr | F444W_mag | F444W_magerr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg,deg | pix2 | pix | pix | deg | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | |||||||||||||||||||||||
| int32 | float64 | float64 | SkyCoord | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 466.24461350033647 | 8.7975642602371 | 53.17381009103326,-27.805320830365776 | 101.0 | 3.121883592723396 | 1.687718238373556 | 0.4593910412587602 | -44.478947493786606 | 0.3750841422808698 | 9.172811357600109 | 0.846427300685747 | 28.9937438445848 | 0.09583064864551505 | 15.251750655416094 | 0.7351865962388724 | 28.441700758604433 | 0.05111393452569058 | 18.35603511311358 | 0.9076155199954206 | 28.240552800545306 | 0.05239928368180803 | 20.355880166244727 | 0.9951286984899145 | 28.128275286137136 | 0.05182128829099082 | 15.305414631812216 | 0.3724290991315014 | 28.437887251597694 | 0.02610307984312319 | 14.02083788279239 | 0.45364676842729956 | 28.533065080581018 | 0.03457285577386587 | 14.174014574304143 | 0.399926108118267 | 28.521267812537104 | 0.030210306449578152 | 13.880611681957355 | 0.46501162852267625 | 28.543978488116917 | 0.035777045316298495 | 13.807578444079855 | 0.4986392632950047 | 28.54970620192208 | 0.038518276199782 |
| 2 | 104.80681791619067 | 18.082129411035602 | 53.17721522973072,-27.805243193448323 | 271.0 | 4.4330760205526705 | 2.8904310038200527 | 0.34798523859744157 | -26.54972210581966 | 0.4292034166747856 | 66.5796776126402 | 2.4362560236234203 | 26.841645780209955 | 0.03901919877231887 | 67.80807336882998 | 1.7819304084598673 | 26.82179648764812 | 0.028163637818553414 | 70.39338414035721 | 2.1096860156757153 | 26.781170389302055 | 0.03206138240515755 | 77.74109040131492 | 2.318681567876357 | 26.6733734305523 | 0.031909302523638994 | 72.90069026757769 | 0.8318824113256227 | 26.743170898753043 | 0.012319367708500414 | 75.27250253951928 | 1.070603105909879 | 26.708409112497115 | 0.015333670289260427 | 71.48792896636 | 0.9342131216075652 | 26.764418211087325 | 0.01409662645806206 | 61.42767389099904 | 0.9993087828855896 | 26.929089824977595 | 0.017520685942298192 | 57.2203760546637 | 1.051093157627436 | 27.00612323080047 | 0.019763151293857934 |
| 3 | 15.04416396551755 | 13.21870964358083 | 53.178060898531776,-27.805283643045666 | 8.0 | 0.8501945455843772 | 0.6682678017840504 | 0.21398248759086114 | -38.1612276642421 | 0.10521135944081007 | 0.3018945818243589 | 0.22003285728443536 | 32.700361703239686 | 0.5943870269087477 | 0.7673344664190465 | 0.20049710017234815 | 31.68753823530937 | 0.25203769246273966 | 0.7185263134982874 | 0.22413743795870192 | 31.758893306827684 | 0.29478532518129674 | 0.9352163612291753 | 0.25774023109081057 | 31.47271975993688 | 0.26428136354091386 | 0.34971362332966227 | 0.06900286982371183 | 32.54071862281815 | 0.19551879280610804 | 0.5607617346802088 | 0.11336299849737179 | 32.02805407366013 | 0.19990472716017532 | 0.26686576820951446 | 0.07056536651518255 | 32.83426782771906 | 0.2547307087052087 | -0.06031654176708369 | 0.08275804453775322 | 99.0 | 34.10547444921151 | 0.11765422427560634 | 0.08200758112663485 | 33.723481188078146 | 0.5742186726193422 |
| 4 | 710.3242889701858 | 14.259777138086877 | 53.17151058505514,-27.805275444028442 | 25.0 | 1.2749753643104456 | 1.117371994707132 | 0.12361287442487268 | -59.26178196559024 | 0.33737600483155716 | 2.656683621955139 | 0.4371488086973456 | 30.339150403981968 | 0.16539237291898956 | 3.818279739887424 | 0.36055789873066885 | 29.945330642292358 | 0.09796938660150925 | 3.9761821672022384 | 0.4246249997227718 | 29.901334316975543 | 0.11016516500461197 | 3.5589744431503423 | 0.42659305165797884 | 30.02168782658959 | 0.12291324714108388 | 1.6962298219026282 | 0.12672768826931047 | 30.826288264009904 | 0.07822962940363362 | 1.590536535091481 | 0.16680910589172168 | 30.896140876425513 | 0.10828384762060109 | 1.9912255148410796 | 0.1511270257305008 | 30.652198878679833 | 0.07942622585637474 | 1.9438353657449134 | 0.178896842573335 | 30.67835130168316 | 0.09558932550457658 | 1.6253103136417588 | 0.17102441486844794 | 30.8726592718879 | 0.1086274373485027 |
| 5 | 434.5294891713508 | 22.219091527997158 | 53.17410887322447,-27.8052089643638 | 268.0 | 3.9551276120227397 | 2.6287674709150584 | 0.33535204706817323 | -8.140060370882344 | 0.5363680123723463 | 48.23502206197299 | 1.7919985963294953 | 27.191593796249776 | 0.039605394744160985 | 54.21551584153409 | 1.365885151142132 | 27.064691014279646 | 0.027014737948708123 | 58.28722242134141 | 1.6235799372401896 | 26.98606659938477 | 0.02982943923746284 | 83.01615938264015 | 2.014897619560197 | 26.602093406179243 | 0.026037350500330014 | 133.10191702875915 | 1.0561372293002846 | 26.089539223653347 | 0.008581100944569356 | 133.48409670470673 | 1.2826536715221066 | 26.086426182655003 | 0.010383073588802694 | 144.0530215352144 | 1.2384723583999508 | 26.003694069729168 | 0.009294542790104875 | 161.5683820059277 | 1.4303041800759473 | 25.879109060386504 | 0.009569320100872941 | 148.33362636536881 | 1.5722468769659783 | 25.971900964480472 | 0.01144758607362489 |
| 6 | 289.38802515014896 | 17.281765562153954 | 53.17547627056455,-27.805250009193262 | 17.0 | 1.2913044931147235 | 1.1978568863866517 | 0.07236682535090477 | 68.12270923149859 | 0.18041073594089005 | 1.1303554119583668 | 0.3821670936376741 | 31.266962455044794 | 0.31621706870061633 | 1.501146258557748 | 0.2867615998533397 | 30.95894247960155 | 0.1898053127105452 | 1.0456239960240308 | 0.28930296167070935 | 31.3515611467238 | 0.2652049050989649 | 1.955256158588051 | 0.38221513653203576 | 30.671990843785842 | 0.1938565595483074 | 1.188616449626284 | 0.1236824441824322 | 31.212395659255414 | 0.10747756611504025 | 1.7708471014756073 | 0.19746701583101794 | 30.77954733764033 | 0.11478335606840087 | 1.492143834211169 | 0.15067991684803003 | 30.965473278295406 | 0.10445071085101643 | 1.1433927169620364 | 0.17563750503987804 | 31.25451144606716 | 0.15514831191999282 | 0.5926553032606401 | 0.13657056678097335 | 31.967994562614585 | 0.2251497308694742 |
| 7 | 640.4863091995317 | 17.773429154661983 | 53.17216853452775,-27.805246129692247 | 21.0 | 1.5124916692884618 | 0.8803106550282112 | 0.41797322067741016 | -64.72678377811528 | 0.257339933672325 | 1.7318562240696873 | 0.35449520565926584 | 30.803720413136418 | 0.20218907261626817 | 1.6434298396482203 | 0.24985582837207415 | 30.86062207958568 | 0.1536624479403839 | 1.6361021041937924 | 0.2840661438634781 | 30.865473992028104 | 0.17382220180983635 | 2.2295861313548446 | 0.33214154219160175 | 30.529439364296703 | 0.15077176392081712 | 1.0399598684843063 | 0.09617790287548726 | 31.357458548945587 | 0.09603604488615963 | 1.6537574517445193 | 0.1507209599380506 | 30.853820464729573 | 0.094699691326289 | 1.2604572151760263 | 0.11591912740405148 | 31.148679728456177 | 0.0955227266471647 | 0.40788132175057795 | 0.11575451167421517 | 32.37366545505972 | 0.27123885135203707 | 0.5688140707837224 | 0.10865005565010251 | 32.01257417244966 | 0.18978993015591245 |
| 8 | 762.762291737285 | 28.289914155884627 | 53.171016553902064,-27.805158549648304 | 200.0 | 3.160315774736445 | 2.3536095208280066 | 0.2552612812799423 | 37.520056617399476 | 0.5417393494467531 | 52.88457336395036 | 1.7905820052498276 | 27.091677487207892 | 0.03615255179943194 | 55.84480419763607 | 1.3404039005228823 | 27.032543068603033 | 0.025752333237818836 | 61.65651418808454 | 1.6299731658033907 | 26.925052580457162 | 0.028330058735061472 | 87.75832428916449 | 2.0319859659923534 | 26.54177919509308 | 0.024852875576127663 | 85.91084431487936 | 0.7872047738101092 | 26.564880031956896 | 0.009903344287860692 | 73.69676407989382 | 0.8932394257702934 | 26.73137895243849 | 0.013080521346957403 | 74.68199206424094 | 0.8363385018198523 | 26.716960265540358 | 0.012091217376307159 | 70.61910087228084 | 0.9001197500128227 | 26.777694540519555 | 0.013751473288938415 | 73.9961654528609 | 1.0387154150809097 | 26.726976963031056 | 0.015134956407915642 |
| 9 | 801.0706722163886 | 32.58026118517374 | 53.17065564474934,-27.805122812753073 | 12.0 | 1.3349434908844906 | 0.6567170220834244 | 0.5080563135685205 | -28.75519616047975 | 0.28793220448127643 | 1.1246036221856506 | 0.3004786448361928 | 31.272501305061585 | 0.2571011450147618 | 1.2797476041233058 | 0.23055260850377246 | 31.132189187101062 | 0.1798473959435111 | 1.2734342201222038 | 0.25959362782849404 | 31.13755870954592 | 0.2014338196131599 | 1.3630620117794083 | 0.2799372864607707 | 31.063710964299986 | 0.20280440914679781 | 1.082787068323561 | 0.10030137862329394 | 31.313642349074232 | 0.0961853826328532 | 0.6781367101230981 | 0.1161012153749803 | 31.821706862430293 | 0.17158341526256418 | 0.5758891042338462 | 0.08989342817468729 | 31.99915284550489 | 0.1574838373578829 | 1.1076176036929608 | 0.13520585441853786 | 31.289025376253583 | 0.12504898101684045 | 1.113433002949251 | 0.14205342023678058 | 31.283339775099364 | 0.1303698266479056 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 318 | 13.420742051379385 | 884.694395861645 | 53.17807529846922,-27.798021345154865 | 185.0 | 3.285450616131066 | 3.147908093244288 | 0.041864127316802446 | -13.393044858704108 | 0.331692826709577 | 18.866842045199054 | 1.965226560149265 | 28.210751966216968 | 0.10758295905106971 | 31.87824391461666 | 1.7141355153754645 | 27.64126402678436 | 0.05686594445019743 | 50.41198352100823 | 2.41110923471416 | 27.143665535948294 | 0.05072509909406772 | 48.62858360693147 | 2.477066617075186 | 27.182770949596527 | 0.05394324520990566 | 36.759914186317175 | 0.8217034694312548 | 27.486563777794004 | 0.024002452426466658 | 33.4590525753193 | 1.007273298718009 | 27.588715901645074 | 0.03220337636185417 | 34.352936437713055 | 0.912840857540001 | 27.56009033543471 | 0.02847398693113529 | 31.658008005086796 | 1.0140664111276134 | 27.648791038562205 | 0.0342328125312021 | 31.811892751708378 | 1.1073440643576584 | 27.643526225913767 | 0.03715062136419812 |
| 319 | 7.027176577444122 | 891.9727894960166 | 53.17813552144662,-27.797960686059632 | 29.0 | 1.603195468602836 | 1.3128724093575075 | 0.18109024440939903 | -82.75208405786572 | 0.254070174248954 | 4.465998147782199 | 0.882646107926147 | 29.775203653457094 | 0.19581293707076908 | 3.5314931272294396 | 0.5906264916500253 | 30.030104086521717 | 0.16790556232616383 | 5.1074722019128656 | 0.7850802429352477 | 29.629484970984358 | 0.15524361227894562 | 4.917640884310548 | 0.8043621780644387 | 29.67060797308068 | 0.16447818783217794 | 3.997287152346768 | 0.2843667768454504 | 29.895586630725628 | 0.07461553548507588 | 3.8377835048269504 | 0.3661371226929502 | 29.93979882029562 | 0.09893508954511895 | 3.412375186414749 | 0.30628802811413675 | 30.067358061850612 | 0.09332518111805055 | 3.28526387152336 | 0.35498246466009514 | 30.10857435571395 | 0.111401289304535 | 3.979156125410199 | 0.40496953684556536 | 29.900522551559803 | 0.10523003616876628 |
| 320 | 143.92969676102126 | 953.6973600108524 | 53.17684577662435,-27.797446433611444 | 59.0 | 2.6507581110195266 | 1.3964068465385502 | 0.47320472557133164 | 25.19703083525805 | 0.2823870637944277 | 5.3423393131722845 | 1.0819792665125496 | 29.580671329181175 | 0.20023900246692783 | 5.922064358315133 | 0.8036616931083888 | 29.46881719301301 | 0.13816512774989942 | 7.212792554437217 | 0.999637029980137 | 29.254741395699362 | 0.1409205427642921 | 13.04894576803854 | 1.3191055611080584 | 28.61106143453415 | 0.10455611158757944 | 8.378792200159028 | 0.41344559288155197 | 29.092046450623155 | 0.052295014155148895 | 6.694537733317119 | 0.5044867944031559 | 29.335698515562246 | 0.07888264867898688 | 6.468848669874378 | 0.4254486120153371 | 29.372932521208615 | 0.06915753681006198 | 7.525370789524617 | 0.5284959208044608 | 29.208680241701657 | 0.07369133634046583 | 6.716742187814203 | 0.5392825802265447 | 29.332103302827207 | 0.08385019406434349 |
| 321 | 142.746173256619 | 959.5389917866399 | 53.176856920479636,-27.797397752374 | 48.0 | 2.1427436610213637 | 1.2555061677914512 | 0.41406609169806174 | -35.47427950196171 | 0.3421221334166607 | 6.936236300337239 | 1.1392607694993395 | 29.29719029986463 | 0.16511345958398996 | 7.903254879897409 | 0.8693971737990356 | 29.15548502960466 | 0.11331229072011467 | 10.187623025402443 | 1.1263006235500606 | 28.87981783423585 | 0.11385094327350063 | 12.948029906646463 | 1.3061016666971015 | 28.619490765397067 | 0.10434267352803422 | 9.023017764882551 | 0.41847771533872685 | 29.011620468937362 | 0.049222442862693484 | 5.447014735335707 | 0.45676894530082573 | 29.559603624665375 | 0.0874297151873417 | 6.472893305711534 | 0.41968922190657787 | 29.37225387876392 | 0.0682088157355045 | 6.488348644754336 | 0.4869395067625267 | 29.369664554504258 | 0.07856993742528491 | 5.76627926777331 | 0.5007325407405385 | 29.497760820098495 | 0.09041210263092743 |
| 322 | 790.415367777511 | 955.3106960545537 | 53.17075559998907,-27.797433388936582 | 93.0 | 2.621691845359079 | 1.8772136125476881 | 0.28396862664438105 | 5.157966396104574 | 0.37596577589838664 | 20.729595554702005 | 1.8703470342379758 | 28.108522927842365 | 0.0937912675982318 | 28.033949382492544 | 1.538051849412483 | 27.78078928786822 | 0.0579910769113244 | 25.743535488305984 | 1.7358437505234157 | 27.873329523763395 | 0.07084681814957344 | 25.620810723619343 | 1.8129005233748652 | 27.87851782980156 | 0.0742292377178118 | 21.15342216887344 | 0.6264899234444314 | 28.086548407739905 | 0.03168871406956517 | 18.828253167515435 | 0.7475941257205683 | 28.212974926843625 | 0.04227634815304789 | 17.96461067150448 | 0.6596358161121416 | 28.26395547593533 | 0.03915225292374026 | 12.790064955174342 | 0.6722158462709531 | 28.632818124807635 | 0.05561473730016507 | 13.580929019180171 | 0.7400945376355254 | 28.567676301555696 | 0.05761144934684235 |
| 323 | 797.3397409211948 | 965.5478941851635 | 53.17069036467991,-27.797348081794517 | 121.0 | 3.0304070507432326 | 1.7633197784086392 | 0.4181244470190332 | 79.51657917516623 | 0.47716929153411286 | 45.619197249141145 | 2.6518036744705875 | 27.25213090293863 | 0.061346663507972285 | 50.392274238753124 | 2.0188186652038733 | 27.144090102966842 | 0.042648142807613955 | 49.831751964789014 | 2.356774844362362 | 27.156234609522436 | 0.05017220365924456 | 48.61650529342564 | 2.4430655896970648 | 27.183040657043588 | 0.05323355846509684 | 33.58368869698832 | 0.7779020039869248 | 27.58467901105726 | 0.024862162100306468 | 23.755213036076142 | 0.8523243778218815 | 27.960602676216816 | 0.03827306144215809 | 23.02386495905596 | 0.7592225343641622 | 27.99455442662465 | 0.03522501052263669 | 19.63315929047223 | 0.8078373356779335 | 28.167524524558377 | 0.043779690921258704 | 18.68731450154886 | 0.8633253572661256 | 28.221132763267338 | 0.049035202457045374 |
| 324 | 790.7983558965504 | 979.5822296300561 | 53.17075198078638,-27.79723112632807 | 133.0 | 4.506451359665357 | 1.493045462994405 | 0.6686871012615841 | -44.19824469199431 | 0.42319913307021234 | 34.20989148182838 | 2.353623899073531 | 27.56462075862663 | 0.07224061886404722 | 36.57135462018657 | 1.7557906133598953 | 27.49214738223006 | 0.05091356319336782 | 42.68849554096421 | 2.206478720252704 | 27.324222876621587 | 0.05471719389692955 | 45.99460251160774 | 2.382028845957703 | 27.243232824978676 | 0.05482188387776275 | 34.80004302822041 | 0.7822981639992332 | 27.54605054768332 | 0.02413684700611631 | 23.40568188262023 | 0.8462317452688508 | 27.976696754900892 | 0.03856178701393752 | 22.76737839931649 | 0.7438457514116649 | 28.006717435787387 | 0.03490554072070799 | 20.501256369918995 | 0.8205498509872132 | 28.120548808604052 | 0.0426087886820206 | 19.089604206836878 | 0.8667809534858215 | 28.198007689828906 | 0.048212383021397735 |
| 325 | 307.7502590912509 | 989.993370703415 | 53.175302486237165,-27.797144093448775 | 37.0 | 1.707727984108737 | 1.1914329585464343 | 0.3023286087519126 | -56.82602073196763 | 0.35854189153102795 | 7.126198163579401 | 1.0919973562362062 | 29.2678552626553 | 0.15479643612317479 | 6.598188007454473 | 0.7664569787646116 | 29.351438284784823 | 0.11931782592149333 | 6.196539377793785 | 0.8658897241714892 | 29.41962696525653 | 0.1420122179341881 | 8.899889611694567 | 1.0602604803697635 | 29.02653845005323 | 0.12220315747396855 | 6.033610248611692 | 0.341615226376123 | 29.44855686812721 | 0.05979574121037518 | 4.942377643470794 | 0.40732286190369604 | 29.66516018385374 | 0.08598385736288833 | 7.336029555914942 | 0.41482302005252747 | 29.23634731904543 | 0.05972101036700092 | 4.176814226515838 | 0.3922167589581281 | 29.84788710139504 | 0.0974473596196291 | 4.604584237652296 | 0.4395488742519671 | 29.742023943768242 | 0.0989902908814326 |
| 326 | 306.8506442261885 | 997.243644725512 | 53.1753109550711,-27.797083673855024 | 51.0 | 2.5021353669231527 | 1.4883885517566502 | 0.4051526662256868 | 15.388098948641728 | 0.24216628412005226 | 8.14019232699493 | 1.1783072479711838 | 29.12341333496192 | 0.14677830930960412 | 10.621922747541108 | 0.9579591476710196 | 28.834492153742637 | 0.09375247870667036 | 10.735153086304802 | 1.1138972395145823 | 28.822979395055036 | 0.1071882551431706 | 12.643670520931202 | 1.2540115652526385 | 28.645317074711215 | 0.10267300679924708 | 9.553009517952841 | 0.41693523286415374 | 28.94964947388319 | 0.04638135299187599 | 9.153235549670372 | 0.5151314172293293 | 28.99606340336343 | 0.05944621805492528 | 10.168036276784187 | 0.47542516670461316 | 28.88190728251713 | 0.04961451062251047 | 7.872874739121797 | 0.5029271829323597 | 29.159666645795102 | 0.06723264138084344 | 7.568902388401964 | 0.5444025773994181 | 29.202417738905766 | 0.07541224065721708 |
Save photometry as output catalog#
tbl.write('JADESphotometry.ecsv', overwrite=True)
!head -175 JADESphotometry.ecsv # show the first 175 lines
# %ECSV 1.0
# ---
# datatype:
# - {name: label, datatype: int32}
# - {name: xcentroid, datatype: float64}
# - {name: ycentroid, datatype: float64}
# - {name: sky_centroid.ra, unit: deg, datatype: float64}
# - {name: sky_centroid.dec, unit: deg, datatype: float64}
# - {name: area, unit: pix2, datatype: float64}
# - {name: a, unit: pix, datatype: float64}
# - {name: b, unit: pix, datatype: float64}
# - {name: ellipticity, datatype: float64}
# - {name: orientation, unit: deg, datatype: float64}
# - {name: gini, datatype: float64}
# - {name: F090W_flux, unit: nJy, datatype: float64}
# - {name: F090W_fluxerr, unit: nJy, datatype: float64}
# - {name: F090W_mag, datatype: float64}
# - {name: F090W_magerr, datatype: float64}
# - {name: F115W_flux, unit: nJy, datatype: float64}
# - {name: F115W_fluxerr, unit: nJy, datatype: float64}
# - {name: F115W_mag, datatype: float64}
# - {name: F115W_magerr, datatype: float64}
# - {name: F150W_flux, unit: nJy, datatype: float64}
# - {name: F150W_fluxerr, unit: nJy, datatype: float64}
# - {name: F150W_mag, datatype: float64}
# - {name: F150W_magerr, datatype: float64}
# - {name: F200W_flux, unit: nJy, datatype: float64}
# - {name: F200W_fluxerr, unit: nJy, datatype: float64}
# - {name: F200W_mag, datatype: float64}
# - {name: F200W_magerr, datatype: float64}
# - {name: F277W_flux, unit: nJy, datatype: float64}
# - {name: F277W_fluxerr, unit: nJy, datatype: float64}
# - {name: F277W_mag, datatype: float64}
# - {name: F277W_magerr, datatype: float64}
# - {name: F335M_flux, unit: nJy, datatype: float64}
# - {name: F335M_fluxerr, unit: nJy, datatype: float64}
# - {name: F335M_mag, datatype: float64}
# - {name: F335M_magerr, datatype: float64}
# - {name: F356W_flux, unit: nJy, datatype: float64}
# - {name: F356W_fluxerr, unit: nJy, datatype: float64}
# - {name: F356W_mag, datatype: float64}
# - {name: F356W_magerr, datatype: float64}
# - {name: F410M_flux, unit: nJy, datatype: float64}
# - {name: F410M_fluxerr, unit: nJy, datatype: float64}
# - {name: F410M_mag, datatype: float64}
# - {name: F410M_magerr, datatype: float64}
# - {name: F444W_flux, unit: nJy, datatype: float64}
# - {name: F444W_fluxerr, unit: nJy, datatype: float64}
# - {name: F444W_mag, datatype: float64}
# - {name: F444W_magerr, datatype: float64}
# meta: !!omap
# - {date: '2025-07-15 18:19:45 UTC'}
# - version: {Python: 3.11.13, astropy: 7.1.0, bottleneck: null, gwcs: 0.25.1, matplotlib: 3.10.3, numpy: 2.3.1, photutils: 2.2.0, scipy: 1.16.0,
# skimage: 0.25.2}
# - {localbkg_width: 0}
# - {apermask_method: correct}
# - kron_params: !!python/tuple [2.5, 1.4, 0.0]
# - __serialized_columns__:
# F090W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: &id001 !astropy.units.Unit {unit: nJy}
# value: !astropy.table.SerializedColumn {name: F090W_flux}
# F090W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F090W_fluxerr}
# F115W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F115W_flux}
# F115W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F115W_fluxerr}
# F150W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F150W_flux}
# F150W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F150W_fluxerr}
# F200W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F200W_flux}
# F200W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F200W_fluxerr}
# F277W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F277W_flux}
# F277W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F277W_fluxerr}
# F335M_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F335M_flux}
# F335M_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F335M_fluxerr}
# F356W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F356W_flux}
# F356W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F356W_fluxerr}
# F410M_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F410M_flux}
# F410M_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F410M_fluxerr}
# F444W_flux:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F444W_flux}
# F444W_fluxerr:
# __class__: astropy.units.quantity.Quantity
# unit: *id001
# value: !astropy.table.SerializedColumn {name: F444W_fluxerr}
# a:
# __class__: astropy.units.quantity.Quantity
# unit: !astropy.units.Unit {unit: pix}
# value: !astropy.table.SerializedColumn {name: a}
# area:
# __class__: astropy.units.quantity.Quantity
# unit: !astropy.units.Unit {unit: pix2}
# value: !astropy.table.SerializedColumn {name: area}
# b:
# __class__: astropy.units.quantity.Quantity
# unit: !astropy.units.Unit {unit: pix}
# value: !astropy.table.SerializedColumn {name: b}
# ellipticity:
# __class__: astropy.units.quantity.Quantity
# unit: !astropy.units.Unit {unit: ''}
# value: !astropy.table.SerializedColumn {name: ellipticity}
# orientation:
# __class__: astropy.units.quantity.Quantity
# unit: &id002 !astropy.units.Unit {unit: deg}
# value: !astropy.table.SerializedColumn {name: orientation}
# sky_centroid:
# __class__: astropy.coordinates.sky_coordinate.SkyCoord
# dec: !astropy.table.SerializedColumn
# __class__: astropy.coordinates.angles.core.Latitude
# unit: *id002
# value: !astropy.table.SerializedColumn {name: sky_centroid.dec}
# frame: icrs
# ra: !astropy.table.SerializedColumn
# __class__: astropy.coordinates.angles.core.Longitude
# unit: *id002
# value: !astropy.table.SerializedColumn {name: sky_centroid.ra}
# wrap_angle: !astropy.coordinates.Angle
# unit: *id002
# value: 360.0
# representation_type: spherical
# schema: astropy-2.0
label xcentroid ycentroid sky_centroid.ra sky_centroid.dec area a b ellipticity orientation gini F090W_flux F090W_fluxerr F090W_mag F090W_magerr F115W_flux F115W_fluxerr F115W_mag F115W_magerr F150W_flux F150W_fluxerr F150W_mag F150W_magerr F200W_flux F200W_fluxerr F200W_mag F200W_magerr F277W_flux F277W_fluxerr F277W_mag F277W_magerr F335M_flux F335M_fluxerr F335M_mag F335M_magerr F356W_flux F356W_fluxerr F356W_mag F356W_magerr F410M_flux F410M_fluxerr F410M_mag F410M_magerr F444W_flux F444W_fluxerr F444W_mag F444W_magerr
1 466.24461350033647 8.7975642602371 53.17381009103326 -27.805320830365776 101.0 3.121883592723396 1.687718238373556 0.4593910412587602 -44.478947493786606 0.3750841422808698 9.172811357600109 0.846427300685747 28.9937438445848 0.09583064864551505 15.251750655416094 0.7351865962388724 28.441700758604433 0.05111393452569058 18.35603511311358 0.9076155199954206 28.240552800545306 0.05239928368180803 20.355880166244727 0.9951286984899145 28.128275286137136 0.05182128829099082 15.305414631812216 0.3724290991315014 28.437887251597694 0.02610307984312319 14.02083788279239 0.45364676842729956 28.533065080581018 0.03457285577386587 14.174014574304143 0.399926108118267 28.521267812537104 0.030210306449578152 13.880611681957355 0.46501162852267625 28.543978488116917 0.035777045316298495 13.807578444079855 0.4986392632950047 28.54970620192208 0.038518276199782
2 104.80681791619067 18.082129411035602 53.17721522973072 -27.805243193448323 271.0 4.4330760205526705 2.8904310038200527 0.34798523859744157 -26.54972210581966 0.4292034166747856 66.5796776126402 2.4362560236234203 26.841645780209955 0.03901919877231887 67.80807336882998 1.7819304084598673 26.82179648764812 0.028163637818553414 70.39338414035721 2.1096860156757153 26.781170389302055 0.03206138240515755 77.74109040131492 2.318681567876357 26.6733734305523 0.031909302523638994 72.90069026757769 0.8318824113256227 26.743170898753043 0.012319367708500414 75.27250253951928 1.070603105909879 26.708409112497115 0.015333670289260427 71.48792896636 0.9342131216075652 26.764418211087325 0.01409662645806206 61.42767389099904 0.9993087828855896 26.929089824977595 0.017520685942298192 57.2203760546637 1.051093157627436 27.00612323080047 0.019763151293857934
3 15.04416396551755 13.21870964358083 53.178060898531776 -27.805283643045666 8.0 0.8501945455843772 0.6682678017840504 0.21398248759086114 -38.1612276642421 0.10521135944081007 0.3018945818243589 0.22003285728443536 32.700361703239686 0.5943870269087477 0.7673344664190465 0.20049710017234815 31.68753823530937 0.25203769246273966 0.7185263134982874 0.22413743795870192 31.758893306827684 0.29478532518129674 0.9352163612291753 0.25774023109081057 31.47271975993688 0.26428136354091386 0.34971362332966227 0.06900286982371183 32.54071862281815 0.19551879280610804 0.5607617346802088 0.11336299849737179 32.02805407366013 0.19990472716017532 0.26686576820951446 0.07056536651518255 32.83426782771906 0.2547307087052087 -0.06031654176708369 0.08275804453775322 99.0 34.10547444921151 0.11765422427560634 0.08200758112663485 33.723481188078146 0.5742186726193422
4 710.3242889701858 14.259777138086877 53.17151058505514 -27.805275444028442 25.0 1.2749753643104456 1.117371994707132 0.12361287442487268 -59.26178196559024 0.33737600483155716 2.656683621955139 0.4371488086973456 30.339150403981968 0.16539237291898956 3.818279739887424 0.36055789873066885 29.945330642292358 0.09796938660150925 3.9761821672022384 0.4246249997227718 29.901334316975543 0.11016516500461197 3.5589744431503423 0.42659305165797884 30.02168782658959 0.12291324714108388 1.6962298219026282 0.12672768826931047 30.826288264009904 0.07822962940363362 1.590536535091481 0.16680910589172168 30.896140876425513 0.10828384762060109 1.9912255148410796 0.1511270257305008 30.652198878679833 0.07942622585637474 1.9438353657449134 0.178896842573335 30.67835130168316 0.09558932550457658 1.6253103136417588 0.17102441486844794 30.8726592718879 0.1086274373485027
5 434.5294891713508 22.219091527997158 53.17410887322447 -27.8052089643638 268.0 3.9551276120227397 2.6287674709150584 0.33535204706817323 -8.140060370882344 0.5363680123723463 48.23502206197299 1.7919985963294953 27.191593796249776 0.039605394744160985 54.21551584153409 1.365885151142132 27.064691014279646 0.027014737948708123 58.28722242134141 1.6235799372401896 26.98606659938477 0.02982943923746284 83.01615938264015 2.014897619560197 26.602093406179243 0.026037350500330014 133.10191702875915 1.0561372293002846 26.089539223653347 0.008581100944569356 133.48409670470673 1.2826536715221066 26.086426182655003 0.010383073588802694 144.0530215352144 1.2384723583999508 26.003694069729168 0.009294542790104875 161.5683820059277 1.4303041800759473 25.879109060386504 0.009569320100872941 148.33362636536881 1.5722468769659783 25.971900964480472 0.01144758607362489
6 289.38802515014896 17.281765562153954 53.17547627056455 -27.805250009193262 17.0 1.2913044931147235 1.1978568863866517 0.07236682535090477 68.12270923149859 0.18041073594089005 1.1303554119583668 0.3821670936376741 31.266962455044794 0.31621706870061633 1.501146258557748 0.2867615998533397 30.95894247960155 0.1898053127105452 1.0456239960240308 0.28930296167070935 31.3515611467238 0.2652049050989649 1.955256158588051 0.38221513653203576 30.671990843785842 0.1938565595483074 1.188616449626284 0.1236824441824322 31.212395659255414 0.10747756611504025 1.7708471014756073 0.19746701583101794 30.77954733764033 0.11478335606840087 1.492143834211169 0.15067991684803003 30.965473278295406 0.10445071085101643 1.1433927169620364 0.17563750503987804 31.25451144606716 0.15514831191999282 0.5926553032606401 0.13657056678097335 31.967994562614585 0.2251497308694742
7 640.4863091995317 17.773429154661983 53.17216853452775 -27.805246129692247 21.0 1.5124916692884618 0.8803106550282112 0.41797322067741016 -64.72678377811528 0.257339933672325 1.7318562240696873 0.35449520565926584 30.803720413136418 0.20218907261626817 1.6434298396482203 0.24985582837207415 30.86062207958568 0.1536624479403839 1.6361021041937924 0.2840661438634781 30.865473992028104 0.17382220180983635 2.2295861313548446 0.33214154219160175 30.529439364296703 0.15077176392081712 1.0399598684843063 0.09617790287548726 31.357458548945587 0.09603604488615963 1.6537574517445193 0.1507209599380506 30.853820464729573 0.094699691326289 1.2604572151760263 0.11591912740405148 31.148679728456177 0.0955227266471647 0.40788132175057795 0.11575451167421517 32.37366545505972 0.27123885135203707 0.5688140707837224 0.10865005565010251 32.01257417244966 0.18978993015591245
8 762.762291737285 28.289914155884627 53.171016553902064 -27.805158549648304 200.0 3.160315774736445 2.3536095208280066 0.2552612812799423 37.520056617399476 0.5417393494467531 52.88457336395036 1.7905820052498276 27.091677487207892 0.03615255179943194 55.84480419763607 1.3404039005228823 27.032543068603033 0.025752333237818836 61.65651418808454 1.6299731658033907 26.925052580457162 0.028330058735061472 87.75832428916449 2.0319859659923534 26.54177919509308 0.024852875576127663 85.91084431487936 0.7872047738101092 26.564880031956896 0.009903344287860692 73.69676407989382 0.8932394257702934 26.73137895243849 0.013080521346957403 74.68199206424094 0.8363385018198523 26.716960265540358 0.012091217376307159 70.61910087228084 0.9001197500128227 26.777694540519555 0.013751473288938415 73.9961654528609 1.0387154150809097 26.726976963031056 0.015134956407915642
Reformat output catalog for readability (optional)#
# Remove units (pixels) from area
tbl['area'] = tbl['area'].value.astype(int)
# Replace sky_centroid with ra, dec
tbl['ra'] = tbl['sky_centroid'].ra.degree
tbl['dec'] = tbl['sky_centroid'].dec.degree
columns = list(tbl.columns)
columns = columns[:3] + ['ra', 'dec'] + columns[4:-2]
tbl = tbl[columns]
for column in columns:
tbl[column].info.format = '.4f'
tbl['ra'].info.format = '11.7f'
tbl['dec'].info.format = '11.7f'
tbl['label'].info.format = 'd'
tbl['area'].info.format = 'd'
tbl.write('JADESphotometry.cat', format='ascii.fixed_width_two_line', delimiter=' ', overwrite=True)
!head -10 JADESphotometry.cat # show the first 10 lines
label xcentroid ycentroid ra dec area a b ellipticity orientation gini F090W_flux F090W_fluxerr F090W_mag F090W_magerr F115W_flux F115W_fluxerr F115W_mag F115W_magerr F150W_flux F150W_fluxerr F150W_mag F150W_magerr F200W_flux F200W_fluxerr F200W_mag F200W_magerr F277W_flux F277W_fluxerr F277W_mag F277W_magerr F335M_flux F335M_fluxerr F335M_mag F335M_magerr F356W_flux F356W_fluxerr F356W_mag F356W_magerr F410M_flux F410M_fluxerr F410M_mag F410M_magerr F444W_flux F444W_fluxerr F444W_mag F444W_magerr
----- --------- --------- ----------- ----------- ---- ------- ------ ----------- ----------- ------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------ ---------- ------------- --------- ------------
1 466.2446 8.7976 53.1738101 -27.8053208 101 3.1219 1.6877 0.4594 -44.4789 0.3751 9.1728 0.8464 28.9937 0.0958 15.2518 0.7352 28.4417 0.0511 18.3560 0.9076 28.2406 0.0524 20.3559 0.9951 28.1283 0.0518 15.3054 0.3724 28.4379 0.0261 14.0208 0.4536 28.5331 0.0346 14.1740 0.3999 28.5213 0.0302 13.8806 0.4650 28.5440 0.0358 13.8076 0.4986 28.5497 0.0385
2 104.8068 18.0821 53.1772152 -27.8052432 271 4.4331 2.8904 0.3480 -26.5497 0.4292 66.5797 2.4363 26.8416 0.0390 67.8081 1.7819 26.8218 0.0282 70.3934 2.1097 26.7812 0.0321 77.7411 2.3187 26.6734 0.0319 72.9007 0.8319 26.7432 0.0123 75.2725 1.0706 26.7084 0.0153 71.4879 0.9342 26.7644 0.0141 61.4277 0.9993 26.9291 0.0175 57.2204 1.0511 27.0061 0.0198
3 15.0442 13.2187 53.1780609 -27.8052836 8 0.8502 0.6683 0.2140 -38.1612 0.1052 0.3019 0.2200 32.7004 0.5944 0.7673 0.2005 31.6875 0.2520 0.7185 0.2241 31.7589 0.2948 0.9352 0.2577 31.4727 0.2643 0.3497 0.0690 32.5407 0.1955 0.5608 0.1134 32.0281 0.1999 0.2669 0.0706 32.8343 0.2547 -0.0603 0.0828 99.0000 34.1055 0.1177 0.0820 33.7235 0.5742
4 710.3243 14.2598 53.1715106 -27.8052754 25 1.2750 1.1174 0.1236 -59.2618 0.3374 2.6567 0.4371 30.3392 0.1654 3.8183 0.3606 29.9453 0.0980 3.9762 0.4246 29.9013 0.1102 3.5590 0.4266 30.0217 0.1229 1.6962 0.1267 30.8263 0.0782 1.5905 0.1668 30.8961 0.1083 1.9912 0.1511 30.6522 0.0794 1.9438 0.1789 30.6784 0.0956 1.6253 0.1710 30.8727 0.1086
5 434.5295 22.2191 53.1741089 -27.8052090 268 3.9551 2.6288 0.3354 -8.1401 0.5364 48.2350 1.7920 27.1916 0.0396 54.2155 1.3659 27.0647 0.0270 58.2872 1.6236 26.9861 0.0298 83.0162 2.0149 26.6021 0.0260 133.1019 1.0561 26.0895 0.0086 133.4841 1.2827 26.0864 0.0104 144.0530 1.2385 26.0037 0.0093 161.5684 1.4303 25.8791 0.0096 148.3336 1.5722 25.9719 0.0114
6 289.3880 17.2818 53.1754763 -27.8052500 17 1.2913 1.1979 0.0724 68.1227 0.1804 1.1304 0.3822 31.2670 0.3162 1.5011 0.2868 30.9589 0.1898 1.0456 0.2893 31.3516 0.2652 1.9553 0.3822 30.6720 0.1939 1.1886 0.1237 31.2124 0.1075 1.7708 0.1975 30.7795 0.1148 1.4921 0.1507 30.9655 0.1045 1.1434 0.1756 31.2545 0.1551 0.5927 0.1366 31.9680 0.2251
7 640.4863 17.7734 53.1721685 -27.8052461 21 1.5125 0.8803 0.4180 -64.7268 0.2573 1.7319 0.3545 30.8037 0.2022 1.6434 0.2499 30.8606 0.1537 1.6361 0.2841 30.8655 0.1738 2.2296 0.3321 30.5294 0.1508 1.0400 0.0962 31.3575 0.0960 1.6538 0.1507 30.8538 0.0947 1.2605 0.1159 31.1487 0.0955 0.4079 0.1158 32.3737 0.2712 0.5688 0.1087 32.0126 0.1898
8 762.7623 28.2899 53.1710166 -27.8051585 200 3.1603 2.3536 0.2553 37.5201 0.5417 52.8846 1.7906 27.0917 0.0362 55.8448 1.3404 27.0325 0.0258 61.6565 1.6300 26.9251 0.0283 87.7583 2.0320 26.5418 0.0249 85.9108 0.7872 26.5649 0.0099 73.6968 0.8932 26.7314 0.0131 74.6820 0.8363 26.7170 0.0121 70.6191 0.9001 26.7777 0.0138 73.9962 1.0387 26.7270 0.0151
Start new session and analyze results#
Load catalog and segmentation map#
# Catalog: ecsv format preserves units for loading in Python notebooks
tbl = QTable.read('JADESphotometry.ecsv')
# Reconstitute filter list
filters = []
for param in tbl.columns:
if param[-4:] == '_mag':
filters.append(param[:-4])
# Segmentation map
segmfile = 'JADES_detections_segm.fits'
segm = fits.open(segmfile)[0].data
segm = SegmentationImage(segm)
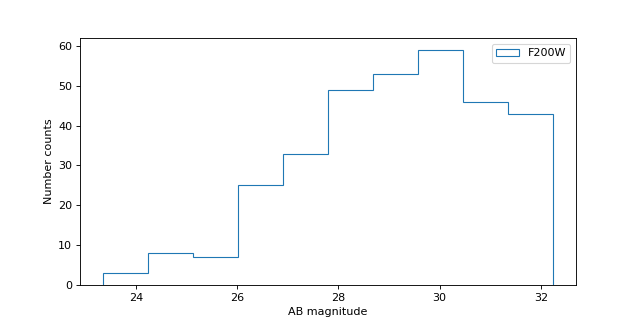
Plot number counts vs. magnitude#
fig = plt.figure(figsize=(8, 4))
filt = 'F200W'
mag1 = tbl[filt + '_mag']
mag1 = mag1[(0 < mag1) & (mag1 < 90)] # detections only
n = plt.hist(mag1, histtype='step', label=filt)
plt.xlabel('AB magnitude')
plt.ylabel('Number counts')
plt.legend()
<matplotlib.legend.Legend at 0x7f0694867190>
Plot F200W vs. F090W magnitudes and look for dropouts#
#import mplcursors
# Would love a better solution here!
mag1 = tbl['F090W_mag']
mag2 = tbl['F200W_mag']
# Only plot detections in F200W
det2 = (0 < mag2) & (mag2 < 90)
mag1 = mag1[det2]
mag2 = mag2[det2]
ids = tbl['label'][det2]
plt.figure(figsize=(8, 4))
plt.plot(mag1, mag2, '.')
for i in range(len(mag1)):
plt.text(mag1[i], mag2[i], ids[i])
plt.xlabel('F090W AB magnitude')
plt.ylabel('F200W AB magnitude')
Text(0, 0.5, 'F200W AB magnitude')
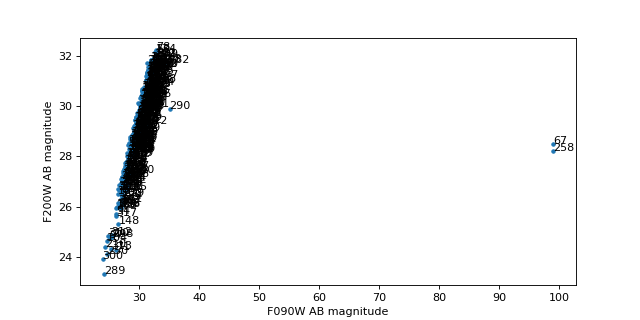
Look at one object#
# Could select object by position
#x, y = 905, 276
#id = segm.data[y,x]
# Select by ID number
id = 261 # F090W dropout
obj = tbl[id-1]
obj
| label | xcentroid | ycentroid | sky_centroid | area | a | b | ellipticity | orientation | gini | F090W_flux | F090W_fluxerr | F090W_mag | F090W_magerr | F115W_flux | F115W_fluxerr | F115W_mag | F115W_magerr | F150W_flux | F150W_fluxerr | F150W_mag | F150W_magerr | F200W_flux | F200W_fluxerr | F200W_mag | F200W_magerr | F277W_flux | F277W_fluxerr | F277W_mag | F277W_magerr | F335M_flux | F335M_fluxerr | F335M_mag | F335M_magerr | F356W_flux | F356W_fluxerr | F356W_mag | F356W_magerr | F410M_flux | F410M_fluxerr | F410M_mag | F410M_magerr | F444W_flux | F444W_fluxerr | F444W_mag | F444W_magerr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg,deg | pix2 | pix | pix | deg | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | nJy | |||||||||||||||||||||||
| int32 | float64 | float64 | SkyCoord | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
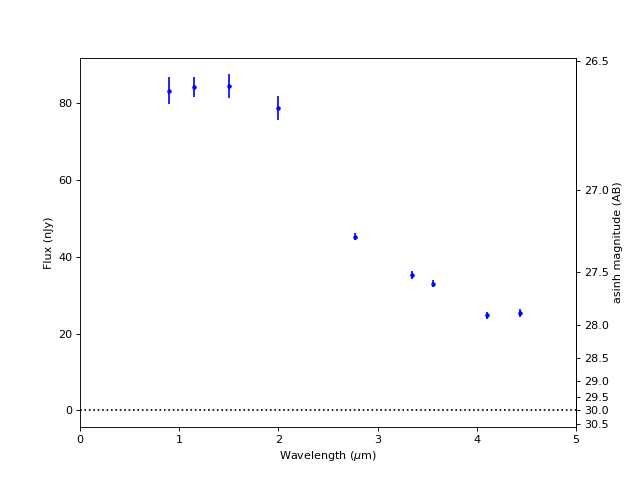
| 261 | 713.6003231522444 | 868.2862731469344 | 53.17147927632734,-27.79815855888541 | 111.0 | 2.1957364237904744 | 1.871859861852015 | 0.14750247726881305 | -17.338774337937068 | 0.546245283985238 | 83.210645372822 | 3.527254135588318 | 26.599552774088977 | 0.04507502616775405 | 84.18846432688923 | 2.587997491179074 | 26.586868530823118 | 0.032873376920099 | 84.42481646179198 | 3.054886353923348 | 26.583824687102044 | 0.038592934581143754 | 78.728836865202 | 3.131787729879179 | 26.659665411873668 | 0.04235304682595798 | 45.258779395038786 | 0.9397610471453727 | 27.260742907280413 | 0.022313544876442264 | 35.277809352747205 | 1.0051267218729336 | 27.53124597775305 | 0.03050203683042903 | 32.91303372370136 | 0.9126975788091346 | 27.60658021307747 | 0.02969819980754908 | 24.75353531926416 | 0.9197377117713978 | 27.915906915058187 | 0.039610013762955344 | 25.401315400116076 | 1.0402217740639617 | 27.887859482444238 | 0.043576230399079735 |
obj['ellipticity']
segmobj = segm.segments[segm.get_index(id)]
segmobj
<photutils.segmentation.core.Segment>
label: 261
slices: (slice(863, 874, None), slice(708, 720, None))
area: 111
Show the object in all the images#
fig, ax = plt.subplots(2, len(filters)+1, figsize=(9.5, 3.5), sharex=True, sharey=True)
ax[0, 0].imshow(rgb[segmobj.slices], origin='lower', extent=segmobj.bbox.extent)
ax[0, 0].set_title('Color')
cmap = segm.make_cmap(seed=12345) # ERROR
ax[1, 0].imshow(segm.data[segmobj.slices], origin='lower', extent=segmobj.bbox.extent, cmap=cmap,
interpolation='nearest')
ax[1, 0].set_title('Segment')
for i in range(1, len(filters)+1):
filt = filters[i-1]
# Show data on top row
data = fits.open(imagefiles[filt])[0].data
stamp = data[segmobj.slices]
norm = ImageNormalize(stretch=SqrtStretch()) # scale each filter individually
ax[0, i].imshow(stamp, extent=segmobj.bbox.extent, cmap='Greys_r', norm=norm, origin='lower')
ax[0, i].set_title(filt.upper())
# Show weights on bottom row
weight = fits.open(weightfiles[filt])[0].data
stamp = weight[segmobj.slices]
# set black to zero weight (no exposure time / bad pixel)
ax[1, i].imshow(stamp, extent=segmobj.bbox.extent, vmin=0, cmap='Greys_r', origin='lower')
ax[0, 0].set_ylabel('Data')
ax[1, 0].set_ylabel('Weight')
Text(0, 0.5, 'Weight')
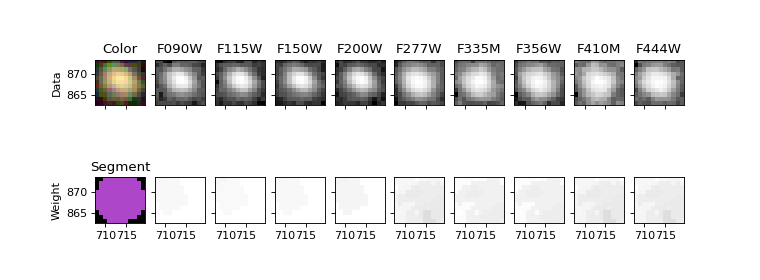
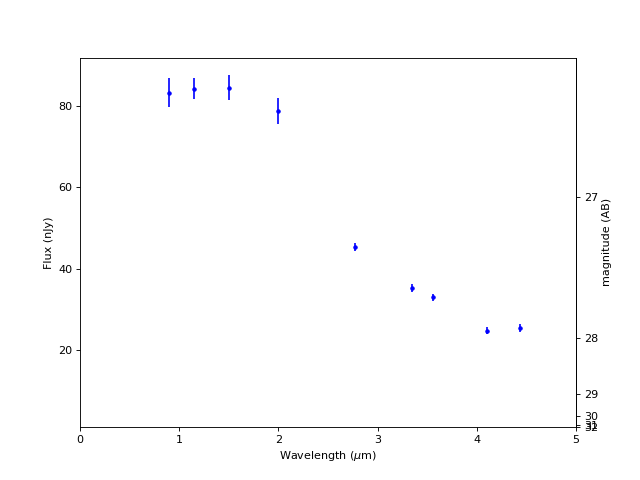
Plot SED (Spectral Energy Distribution)#
fig, ax = plt.subplots(figsize=(8, 6))
for filt in filters:
lam = int(filt[1:4]) / 100
plt.errorbar(lam, obj[filt+'_flux'].value, obj[filt+'_fluxerr'].value, marker='.', c='b')
plt.axhline(0, c='k', ls=':')
plt.xlim(0, 5)
plt.xlabel('Wavelength ($\mu$m)')
plt.ylabel('Flux (nJy)')
mlim = 31.4
flim = mlim * u.ABmag
flim = flim.to(u.nJy).value
# Add AB magnitudes as secondary x-axis at right
# https://matplotlib.org/gallery/subplots_axes_and_figures/secondary_axis.html#sphx-glr-gallery-subplots-axes-and-figures-secondary-axis-py
def AB2nJy(mAB):
m = mAB * u.ABmag
f = m.to(u.nJy)
f = f.value
f = np.where(f > flim, f, flim)
return f
def nJy2AB(F_nJy):
f = F_nJy * u.nJy
m = f.to(u.ABmag)
m = m.value
m = np.where(m < mlim, m, mlim)
return m
plt.ylim(flim, plt.ylim()[1])
secax = ax.secondary_yaxis('right', functions=(nJy2AB, AB2nJy))
secax.set_ylabel('magnitude (AB)')
Text(0, 0.5, 'magnitude (AB)')
Magnitude conversion fails for flux <= 0#
fig, ax = plt.subplots(figsize=(8, 6))
for filt in filters:
lam = int(filt[1:4]) / 100
plt.errorbar(lam, obj[filt+'_flux'].value, obj[filt+'_fluxerr'].value, marker='.', c='b')
plt.axhline(0, c='k', ls=':')
plt.xlim(0, 5)
plt.xlabel('Wavelength ($\mu$m)')
plt.ylabel('Flux (nJy)')
f0 = 10**(0.4 * 31.4) # flux [nJy] at zero magnitude
b0 = 1.e-12 # this should be filter dependent
# Add AB magnitudes as secondary x-axis at right
# https://matplotlib.org/gallery/subplots_axes_and_figures/secondary_axis.html#sphx-glr-gallery-subplots-axes-and-figures-secondary-axis-py
def AB2nJy(m):
f = np.sinh(-0.4 * m * np.log(10) - np.log(b0)) * 2 * b0 * f0
return f
# Luptitudes
# https://www.sdss.org/dr12/algorithms/magnitudes/
def nJy2AB(f):
m = -2.5 / np.log(10) * (np.arcsinh((f / f0) / (2 * b0)) + np.log(b0))
return m
#plt.ylim(flim, plt.ylim()[1])
secax = ax.secondary_yaxis('right', functions=(nJy2AB, AB2nJy))
secax.set_ylabel('asinh magnitude (AB)')
Text(0, 0.5, 'asinh magnitude (AB)')