Improving Accuracy of World Coordinate System of Pure Parallel Dataset#
Introduction#
In this notebook, we will go through the steps needed to improve the accuracy of the World Coordinate System (WCS) in the headers of pure parallel datasets that were observed with JWST prior to the installation of DMS 10.2 (April 2024). The example dataset is jw01571078001_03201_00001_nis_rate.fits, which is a direct image from pure parallel Program ID 1571 (PI: Malkan). Note that the update_parallel_wcs.py script has to be run on all individual datasets taken during a pure parallel visit in order to get the improved WCS for that visit.
In this notebook, we assume that all relevant files are located in the current working directory.
Install pipeline and other required packages#
The required packages are in the provided file requirements.txt. We generally recommend to create a fresh conda environment followed by the installation of those required packages:
conda create -n improve_pure_parallel_wcs
conda activate improve_pure_parallel_wcs
pip install -r requirements.txt
Date last published: February 4, 2025
Imports#
The imports in the next cell are needed to run this notebook.
import copy
import jwst.datamodels as dm
import numpy as np
from astropy.io import ascii
from astropy import units as u
from astropy.coordinates import SkyCoord
from astropy.table import Table
from astropy.time import Time
from astropy.convolution import interpolate_replace_nans, Gaussian2DKernel
import astropy.io.fits as pyfits
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import pysiaf
import warnings
import urllib.request
from astroquery.gaia import Gaia
%matplotlib inline
Define a few functions#
# This example is to show the (mis-)alignment of the World Coordinate System of a Pure Parallel imaging dataset
# taken before the installation of DMS 10.2, relative to GAIA DR3, and how the application of the
# "update_parallel_wcs.py" script improves that alignment.
#
# First some functions to obtain a catalog of GAIA stars near a certain sky position:
#
def query_gaia(ra, dec, radius, verbose=False, epoch=None, filename=None):
"""
Execute a Gaia DR3 query using astroquery, return the table of sources.
Parameters
----------
ra: a float value, the right ascension in decimal degrees
dec: a float value, the declination in decimal degrees
radius: an optional float value, the search radius in arc-seconds,
default 1.0
epoch: an optional float value, the epoch for the positions in decimal
years (2023.197588611 for example, for 2021-05-24T17:46:12.814)
if given, the proper motion is allowed for in the positions that
are returned
verbose: an optional boolean value, if True print a list of the sources
filename: an optional string value, used as the output file name if
the verbose flag is set (if None, print only to the terminal)
Returns
-------
gaiadata1: an Astropy Table containing the catalogue of sources
"""
coord = SkyCoord(ra=ra, dec=dec, unit=(u.degree, u.degree),
frame='icrs')
radius = u.Quantity(radius/3600.0, u.deg)
Gaia.ROW_LIMIT = -1
Gaia.MAIN_GAIA_TABLE = "gaiadr3.gaia_source"
gaiadata1 = Gaia.query_object_async(coordinate=coord, width=radius*2, height=radius*2)
if verbose:
gphot = gaiadata1['phot_g_mean_mag']
gbphot = gaiadata1['phot_bp_mean_mag']
grphot = gaiadata1['phot_rp_mean_mag']
parallax = gaiadata1['parallax']
names = gaiadata1['designation']
with warnings.catch_warnings():
warnings.simplefilter("ignore")
gabs = gphot-5.*np.log10(1000./parallax)+5.
gcol = gbphot - grphot
if epoch is None:
epoch = 2016.0
gaiadata2 = apply_precession(gaiadata1, epoch-2016.0)
ra = gaiadata2['ra']
dec = gaiadata2['dec']
with warnings.catch_warnings():
warnings.simplefilter("ignore")
for ind in range(len(ra)):
g_col = gcol[ind] if (gbphot[ind] < 90.) and (grphot[ind] < 90.) else 0.0
line = f"{ra[ind]:12.8f} {dec[ind]:13.8f} {gphot[ind]:10.6f} {gbphot[ind]:10.6f} {grphot[ind]:10.6f} {parallax[ind]:10.3f} {gabs[ind]:10.4f} {g_col:10.4f} '{names[ind]}'"
line = line.replace('nan', '0.0')
if filename:
with open(filename, 'w') as outfile:
print(' RA DEC g gbp grp parallax abs_g gbp_grp GAIA_Designation', file=outfile)
for ind in range(len(ra)):
g_col = gcol[ind] if (gbphot[ind] < 90.) and (grphot[ind] < 90.) else 0.0
line = f"{ra[ind]:12.8f} {dec[ind]:13.8f} {gphot[ind]:10.6f} {gbphot[ind]:10.6f} {grphot[ind]:10.6f} {parallax[ind]:10.3f} {gabs[ind]:10.4f} {g_col:10.4f} '{names[ind]}'"
line = line.replace('nan', '0.0')
print(line, file=outfile)
return gaiadata2
def apply_precession(catalog, deltat):
"""
Apply precession to update a catalog of values. Uses astropy.SkyCoord
apply_space_motion to update the positions.
Parameters:
catalog: a numpy Table type variable with elements 'ra', 'dec', 'pmra',
and 'pmdec' as per the Gaia DR3 catalog; sky coordinates
must be in degrees and the proper motions must be in mas/year
deltat: a float value, the time change in decimal years for the motion
Returns
-------
newcatalog: a copy of catalog with 'newra', 'newdec' elements with the
revised sky positions; if no proper motion data are
available then newra = ra and newdec = dec
"""
newcatalog = copy.deepcopy(catalog)
newcatalog['newra'] = catalog['ra']
newcatalog['newdec'] = catalog['dec']
with warnings.catch_warnings():
warnings.simplefilter("ignore")
sky_coords = SkyCoord(catalog['ra'], catalog['dec'],
unit=(u.deg, u.deg),
pm_ra_cosdec=catalog['pmra'],
pm_dec=catalog['pmdec'],
obstime=Time('2016-01-01 00:00:00.0'))
newpos = sky_coords.apply_space_motion(dt=deltat*u.yr)
newra = newpos.ra.value
newdec = newpos.dec.value
newcatalog['newra'] = newra
newcatalog['newdec'] = newdec
inds = np.isnan(newra)
newcatalog['newra'][inds] = newcatalog['ra'][inds]
newcatalog['newdec'][inds] = newcatalog['dec'][inds]
return newcatalog
Get sky coordinates of a list of GAIA targets around the pointing of the pure parallel image#
# Now get such a list around pointing of NIRISS pure parallel image jw01571078001_03201_00001_nis_rate.fits
# downloaded from the MAST archive.
# Note: files downloaded or created here will be placed in the current working directory.
boxlink = 'https://stsci.box.com/shared/static/ydxn3hhndwup0qr85fuyqro6suufa6fx.fits'
boxfile = 'jw01571078001_03201_00002_nis_rate.fits'
try:
urllib.request.urlretrieve(boxlink, boxfile)
except Exception as e:
print(f"Error downloading file: {e}")
try:
imfile = pyfits.open(boxfile)
hdr1 = imfile[1].header
ra, dec = hdr1['CRVAL1'], hdr1['CRVAL2']
print(f'RA = {ra}, DEC = {dec}')
except Exception as e:
print(f"Error opening FITS file: {e}")
mygaia = query_gaia(ra, dec, 65., verbose=True, epoch=2022.9968, filename='gaiacoords.out')
RA = 150.41048088035956, DEC = 2.4050524643551943
2025-05-15 19:16:18,833 - stpipe - INFO - Query finished.
INFO: Query finished. [astroquery.utils.tap.core]
Convert those sky coordinates to (x, y) in the image according to its WCS, then show them on image#
t = ascii.read('gaiacoords.out')
ra, dec = (t['RA'], t['DEC'])
ratefile = 'jw01571078001_03201_00002_nis_rate.fits'
instr = imfile[0].header['INSTRUME']
aperture = imfile[0].header['APERNAME']
siaf = pysiaf.Siaf(instr)
myaper = siaf[aperture]
mod = dm.open(ratefile)
am = pysiaf.utils.rotations.attitude_matrix(0, 0, mod.meta.pointing.ra_v1,
mod.meta.pointing.dec_v1,
mod.meta.pointing.pa_v3)
myaper.set_attitude_matrix(am)
x, y = myaper.sky_to_sci(ra, dec)
xytab = Table([ra, dec, x, y], names=('RA', 'DEC', 'x', 'y'))
xytab['RA'].info.format = '.8f'
xytab['DEC'].info.format = '.8f'
xytab['x'].info.format = '.4f'
xytab['y'].info.format = '.4f'
print(xytab)
ys, xs = imfile[1].data.shape
fig = plt.figure(figsize=(15, 15))
ax = fig.add_subplot(111)
xsize, ysize = (20, 20)
for i in range(len(t)):
ax.add_patch(patches.Ellipse(
(x[i], y[i]),
(xsize),
(ysize), fill=False, color='red'))
# Create image with bad pixels interpolated over (for display purposes)
kernel = Gaussian2DKernel(x_stddev=2)
fixed_image = interpolate_replace_nans(imfile[1].data, kernel)
ax.imshow(fixed_image, cmap='binary', origin='lower', extent=[0, xs-1, 0, ys-1], vmin=0.7, vmax=2)
RA DEC x y
------------ ---------- --------- ---------
150.41652702 2.39878800 1178.4537 572.5781
150.41591443 2.39797550 1233.7315 582.1378
150.40719349 2.39406168 1646.0505 909.0131
150.42265455 2.40721593 610.8758 484.1616
150.39991658 2.41498142 805.9535 1784.5106
150.42546736 2.41363882 225.3103 506.9972
150.42277119 2.41723805 117.3792 727.9126
150.41347159 2.38794396 1786.8709 450.3341
150.42439347 2.42208304 -160.7871 769.1785
<matplotlib.image.AxesImage at 0x7ffb3aac1910>
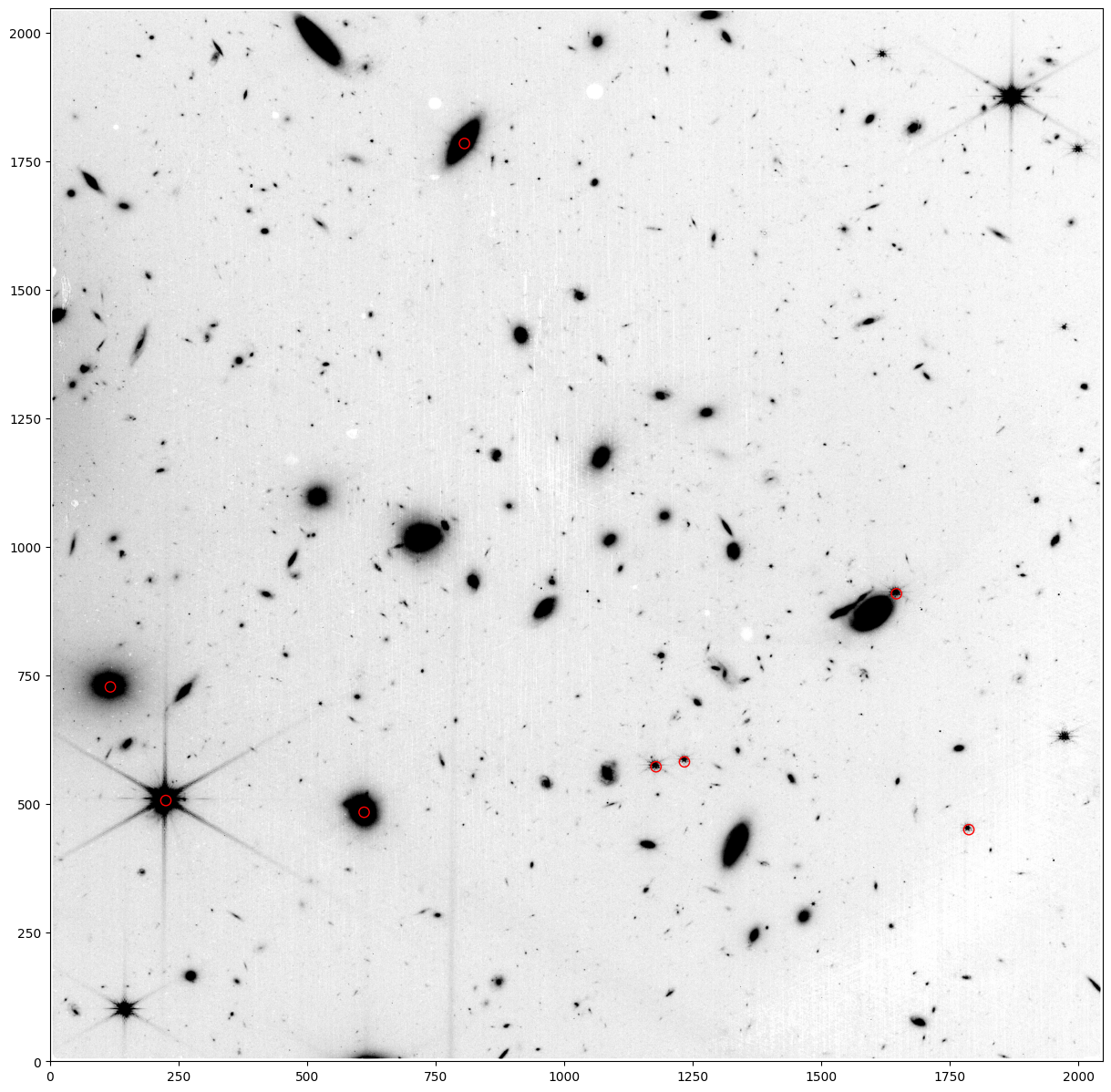
Note that the WCS is ‘off’ by more than a pixel, which is a problem for spectral extraction in the pipeline for WFSS data, since that extraction relies blindly on the accuracy of the WCS.
Now apply the update_parallel_wcs.py script to correct the WCS. Note the offset in pixel coordinates calculated by the script:#
# This cell assumes that you have script update_parallel_wcs.py in your working directory.
# Note: Outside of this notebook, this script should be run as follows on each _rate.fits file in the pure parallel visit:
#
# python update_parallel_wcs.py my_rate.fits <verbosity>
#
# After the script has been run on the input file(s), one can then (re-)run Stage 2 of the JWST calibration pipeline
# (`calwebb_image2` and/or `calwebb_spec2`) on those files, which will now result in correct WCSes in the data headers
# (and corrected locations of spectral extractions in case of pure parallel WFSS data).
#
# By default, `update_parallel_wcs.py` displays the input and output values of the CRVAL1/2 keywords when the script is run
# as seen below. One can avoid this by setting the optional parameter <verbosity> to anything other than `True`.
#
# The script keeps track of its executions using a log file called "pure_parallel_wcs_logfile" in the working directory.
%run update_parallel_wcs.py jw01571078001_03201_00002_nis_rate.fits
PureParallelUtils.update_pure_parallel_wcs: File: jw01571078001_03201_00002_nis_rate.fits
PureParallelUtils.update_pure_parallel_wcs: FGS: jw01762008001_gs-fg_2022364021322_cal.fits
PureParallelUtils.update_pure_parallel_wcs: original crval: 150.4104809 2.4050525
PureParallelUtils.update_pure_parallel_wcs: new crval: 150.4104774 2.4050775
PureParallelUtils.update_pure_parallel_wcs: d(pix): -1.139 0.792
<Figure size 640x480 with 0 Axes>
Calculate (x, y) for the GAIA targets in the updated fits file and print the updated (x, y) coordinates:#
with pyfits.open('jw01571078001_03201_00002_nis_rate.fits') as imfile:
hdr0 = imfile[0].header
myaper = pysiaf.Siaf(hdr0['INSTRUME'])[hdr0['APERNAME']]
mod = dm.open('jw01571078001_03201_00002_nis_rate.fits')
am = pysiaf.utils.rotations.attitude_matrix(0, 0, mod.meta.pointing.ra_v1,
mod.meta.pointing.dec_v1,
mod.meta.pointing.pa_v3)
myaper.set_attitude_matrix(am)
newx, newy = myaper.sky_to_sci(ra, dec)
xytab = Table([x, y, newx, newy], names=('x', 'y', 'x_corr', 'y_corr'))
for col in xytab.colnames:
xytab[col].info.format = '.4f'
print(xytab)
# In practice, one would run the script on all _rate.fits files for a given target,
# then run the stage-2 and stage-3 pipelines on the resulting images
# (specifically, Image2Pipeline for the direct images, followed by Image3Pipeline to create a combined
# image and source catalog, followed by the stage-2 and stage-3 pipelines for the grism images
# (Spec2Pipeline and Spec3Pipeline, respectively).
x y x_corr y_corr
--------- --------- --------- ---------
1178.4537 572.5781 1179.3692 573.1124
1233.7315 582.1378 1234.6468 582.6729
1646.0505 909.0131 1646.9603 909.5544
610.8758 484.1616 611.7927 484.6867
805.9535 1784.5106 806.8472 1785.0388
225.3103 506.9972 226.2269 507.5160
117.3792 727.9126 118.2918 728.4293
1786.8709 450.3341 1787.7893 450.8782
-160.7871 769.1785 -159.8752 769.6907
