Cleaning Residual 1/f Noise in NIRSpec MOS Products with NSClean#
Notebook Goal#
The goal of this notebook is to generate cleaned MOS (_rate.fits) files by removing residual 1/f noise. These cleaned files will be used as input for the level 3 (Spec3Pipeline) pipeline.
Table of Contents#
1. Introduction #
The JWST NIRSpec instrument has a number of features and characteristics that observers should be aware of when planning observations and interpreting data. One notable feature seen in NIRSpec pipeline products is negative and/or surplus flux in the extracted 1-D spectrum, typically with an irregular wavelength-dependent undulation. The cause of this artifact is correlated noise, known as 1/f noise, from low-level detector thermal instabilities, seen as vertical banding in 2-D count rate images, particularly in exposures of the NRS2 detector. While the IRS2 readout mode reduces this effect, it is not completely eliminated.
To address this issue, the JWST Science Calibration Pipeline has integrated an external package developed by Bernard Rauscher, known as NSClean, within the Spec2Pipeline under NSCleanStep. This algorithm uses dark areas of the detector to fit a background model to the data in Fourier space. It requires an input mask to identify all dark areas of the detector. The more thorough and complete this mask is, the better the background fit.
In this notebook, we will use the NSClean algorithm integrated into the pipeline, utilizing a mask generated on-the-fly with default parameters to remove 1/f noise. In some cases, this mask may not be complete enough/too restrictive for the best possible noise removal. To address this, we demonstrate how one can manually modify the default mask, as well as how to create an alternative mask by adjusting the NSCleanStep parameters. If needed, see the NSClean documentation for some suggestions on manually creating a custom mask.
This notebook utilizes a MOS observation from the CEERS program with grating/filter G395M/F290LP, which is part of the JWST Early Release Science program ERS-1345 observation 61, as an example.
2. Import Library #
# ---------- Set CRDS environment variables ----------
import os
import jwst
os.environ['CRDS_CONTEXT'] = 'jwst_1210.pmap'
os.environ['CRDS_PATH'] = os.environ['HOME']+'/crds_cache'
os.environ['CRDS_SERVER_URL'] = 'https://jwst-crds.stsci.edu'
print(f'CRDS cache location: {os.environ["CRDS_PATH"]}')
print("JWST Calibration Pipeline Version={}".format(jwst.__version__))
# print("Current Operational CRDS Context = {}".format(crds.get_default_context()))
CRDS cache location: /home/runner/crds_cache
JWST Calibration Pipeline Version=1.14.0
# ------ General Imports ------
import numpy as np
import time as tt
import logging
import warnings
# ------ JWST Calibration Pipeline Imports ------
from jwst.pipeline.calwebb_spec2 import Spec2Pipeline
# ------ Plotting/Stats Imports ------
from matplotlib import pyplot as plt
from astropy.io import fits
from utils import get_jwst_file, plot_dark_data, plot_cleaned_data, plot_spectra
# Hide all log and warning messages.
logging.disable(logging.ERROR)
warnings.simplefilter("ignore", RuntimeWarning)
3. Download the Data #
The input data for this notebook features a MOS observation from the CEERS program with grating/filter G395H/F290LP. The dataset is part of the JWST Early Release Science program ERS-1345, specifically observation 61. It consists of 3 integrations (3 dither points; 3-SHUTTER-SLITLET) with 9 groups each. This notebook focuses on the third dithered exposure (00003) as an example. However, it’s important to note that before proceeding to the Spec3Pipeline, all exposures must first be processed through the Spec2Pipeline.
# Define a downloads directory.
mast_products_dir = "./mast_products/"
# Check if the directory exists.
if not os.path.exists(mast_products_dir):
# Create the directory if it doesn't exist.
os.makedirs(mast_products_dir)
# This notebook focuses on the third dithered exposure.
obs_ids = ["jw01345061001_07101_00003"]
detectors = [1, 2] # Both Detectors NRS1 and NRS2.
# Specify countrate products
rate_names = []
for obs_id in obs_ids:
for detector in detectors:
rate_names.append(f"{obs_id}_nrs{detector}_rate.fits")
# Download all the FITS files and associated MSA files.
msa_names = []
for name in rate_names:
print(f"Downloading {name}")
get_jwst_file(
name,
mast_api_token="75f915f251544013bb127b7f6f6edc80",
save_directory=mast_products_dir,
)
# Retrieve the MSA file from the header in the downloaded file.
msa_name = fits.getval(mast_products_dir + name, "MSAMETFL")
if not os.path.isfile(msa_name):
print(f"Downloading {msa_name}")
get_jwst_file(
msa_name,
mast_api_token=None,
save_directory=mast_products_dir,
)
msa_names.append(msa_name)
Downloading jw01345061001_07101_00003_nrs1_rate.fits
Downloading jw01345061001_01_msa.fits
Downloading jw01345061001_07101_00003_nrs2_rate.fits
Downloading jw01345061001_01_msa.fits
The file jw01345061001_01_msa.fits already exists in the directory. Skipping download.
4. Running Spec2Pipeline without NSClean (Original Data) #
The cell below executes the Spec2Pipeline, explicitly skipping the NSClean step during processing. The level 2 products generated will serve as a reference point to illustrate how the countrate images and final extracted spectra appear without the removal of 1/f noise.
# Set up directory for running the pipeline without NSClean.
stage2_nsclean_skipped_dir = "./stage2_nsclean_skipped/"
if not os.path.exists(stage2_nsclean_skipped_dir):
os.makedirs(stage2_nsclean_skipped_dir) # Create the directory if it doesn't exist.
# Original data (no NSClean Applied).
# Estimated run-time: 1-2 minutes.
start = tt.time()
for i in rate_names:
print(f"Processing {i}...")
if "nrs1" in i:
slit_name = "80"
else:
slit_name = "11"
Spec2Pipeline.call(
mast_products_dir + i,
save_results=True,
steps={"nsclean": {"skip": True}, "extract_2d": {"slit_name": slit_name}},
output_dir=stage2_nsclean_skipped_dir,
)
print(f"Saved {i[:-9]}" + "cal.fits")
print(f"Saved {i[:-9]}" + "x1d.fits")
end = tt.time()
print("Run time: ", round(end - start, 1) / 60.0, " min")
Processing jw01345061001_07101_00003_nrs1_rate.fits...
Saved jw01345061001_07101_00003_nrs1_cal.fits
Saved jw01345061001_07101_00003_nrs1_x1d.fits
Processing jw01345061001_07101_00003_nrs2_rate.fits...
Saved jw01345061001_07101_00003_nrs2_cal.fits
Saved jw01345061001_07101_00003_nrs2_x1d.fits
Run time: 2.2916666666666665 min
5. Clean up 1/f Noise with NSClean (Default Pipeline Mask) #
If a user-supplied mask file is not provided to the NSClean step in the Spec2Pipeline, the pipeline will generate a mask based on default parameters. This mask will identify any pixel that is unilluminated. That is, the mask must contain True and False values, where True indicates that the pixel is dark, and False indicates that the pixel is illuminated (not dark).
By default, the pipeline marks the following detector areas as illuminated, non-dark areas (False):
Pixels designated as open slits for this MOS observation.
Traces from failed-open MSA shutters.
5-sigma outliers (default value).
Any pixel set to NaN in the rate data.
To tune the outlier detection in the mask, try modifying the n_sigma parameter (explored in the next section). A higher value will identify fewer outliers. A lower value will identify more.
The default generated mask is saved and analyzed below.
# Set up directory for running NSClean with default parameters.
stage2_nsclean_default_dir = "./stage2_nsclean_default/"
if not os.path.exists(stage2_nsclean_default_dir):
os.makedirs(stage2_nsclean_default_dir) # Create the directory if it doesn't exist.
# 1/f noise cleaned data (default NSClean pipeline mask).
# Estimated run time: 6 minutes.
start = tt.time()
for i in rate_names:
print(f"Processing {i}...")
if "nrs1" in i:
slit_name = "80"
else:
slit_name = "11"
Spec2Pipeline.call(
mast_products_dir + i,
save_results=True,
steps={
"nsclean": {"skip": False, "save_mask": True, "save_results": True},
"extract_2d": {"slit_name": slit_name},
},
output_dir=stage2_nsclean_default_dir,
)
print(f"Saved {i[:-9]}" + "mask.fits")
print(f"Saved {i[:-9]}" + "nsclean.fits")
print(f"Saved {i[:-9]}" + "cal.fits")
print(f"Saved {i[:-9]}" + "x1d.fits")
end = tt.time()
print("Run time: ", round(end - start, 1) / 60.0, " min")
Processing jw01345061001_07101_00003_nrs1_rate.fits...
Saved jw01345061001_07101_00003_nrs1_mask.fits
Saved jw01345061001_07101_00003_nrs1_nsclean.fits
Saved jw01345061001_07101_00003_nrs1_cal.fits
Saved jw01345061001_07101_00003_nrs1_x1d.fits
Processing jw01345061001_07101_00003_nrs2_rate.fits...
Saved jw01345061001_07101_00003_nrs2_mask.fits
Saved jw01345061001_07101_00003_nrs2_nsclean.fits
Saved jw01345061001_07101_00003_nrs2_cal.fits
Saved jw01345061001_07101_00003_nrs2_x1d.fits
Run time: 4.196666666666667 min
In some situations, the NSClean step may fail to find a fit to the background noise. This failure may occur if the mask does not contain enough dark data (marked True). In particular, every column in the mask except for the first and last 4 columns must contain some pixels marked True. The background fitting procedure considers each column, one at a time, so it will crash if there is no data in a column to fit. If failure occurs, check that your mask in the image below has at least some True values in every column.
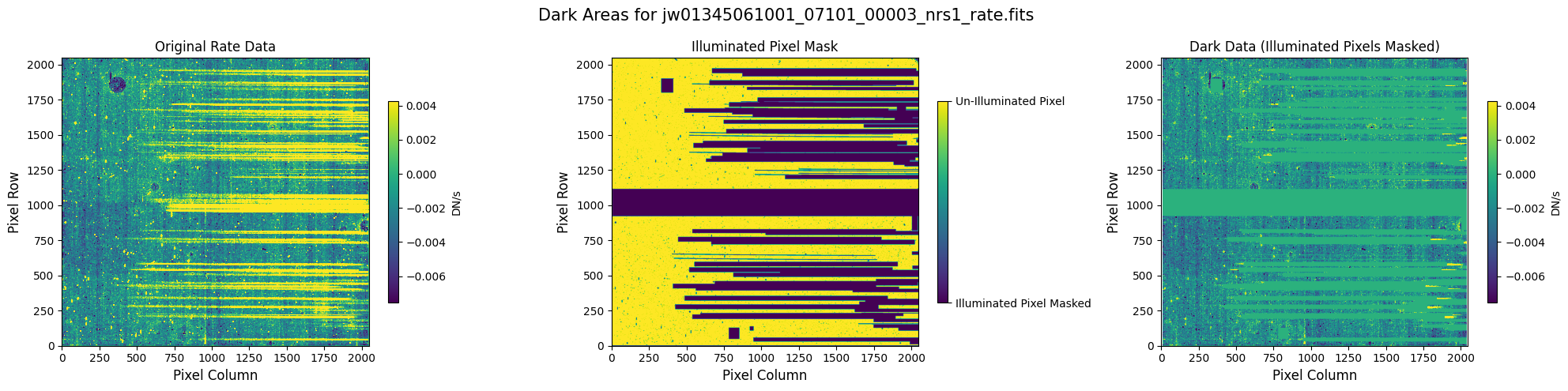
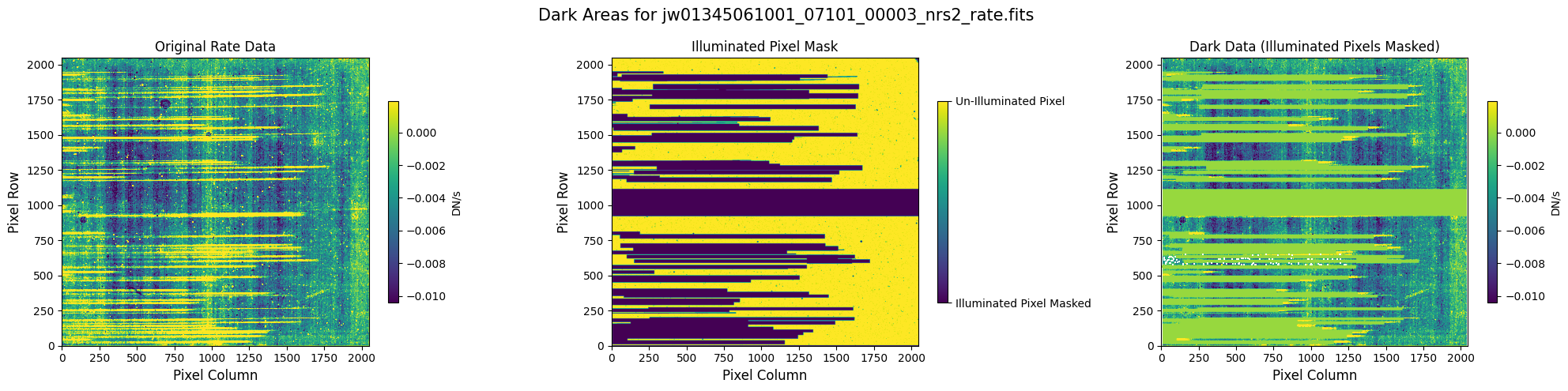
5.1 Verify the Mask (Default Pipeline Mask) #
Check the mask against the rate data to make sure it keeps only dark areas of the detector.
Note that there are still some remaining illuminated areas, primarily due to transient artifacts like cosmic rays and snowballs.
# Plot the rate data with masked areas blocked.
# List of on-the-fly built masks from the pipeline.
nsclean_default_masks = [
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs1_mask.fits",
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs2_mask.fits",
]
# Plot each associated set of rateint data and mask file.
for rate_file, mask_file in zip(rate_names, nsclean_default_masks):
plot_dark_data(mast_products_dir + rate_file, mask_file, layout="columns", scale=9)
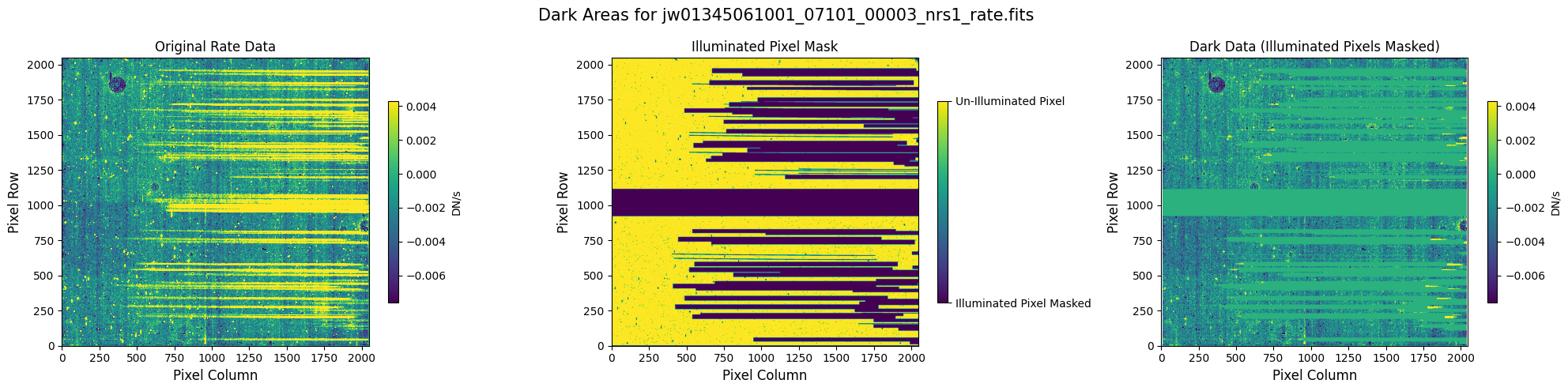
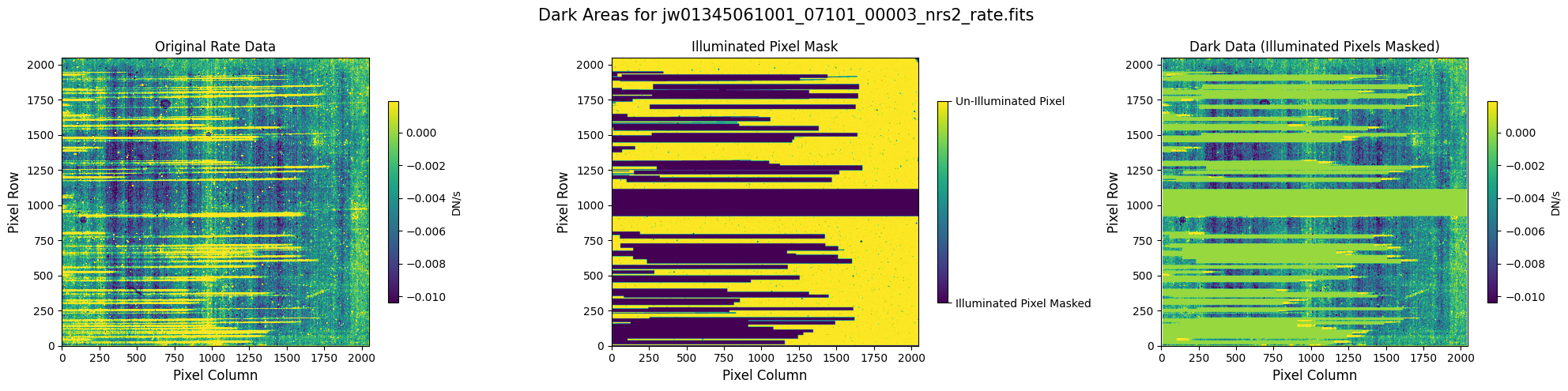
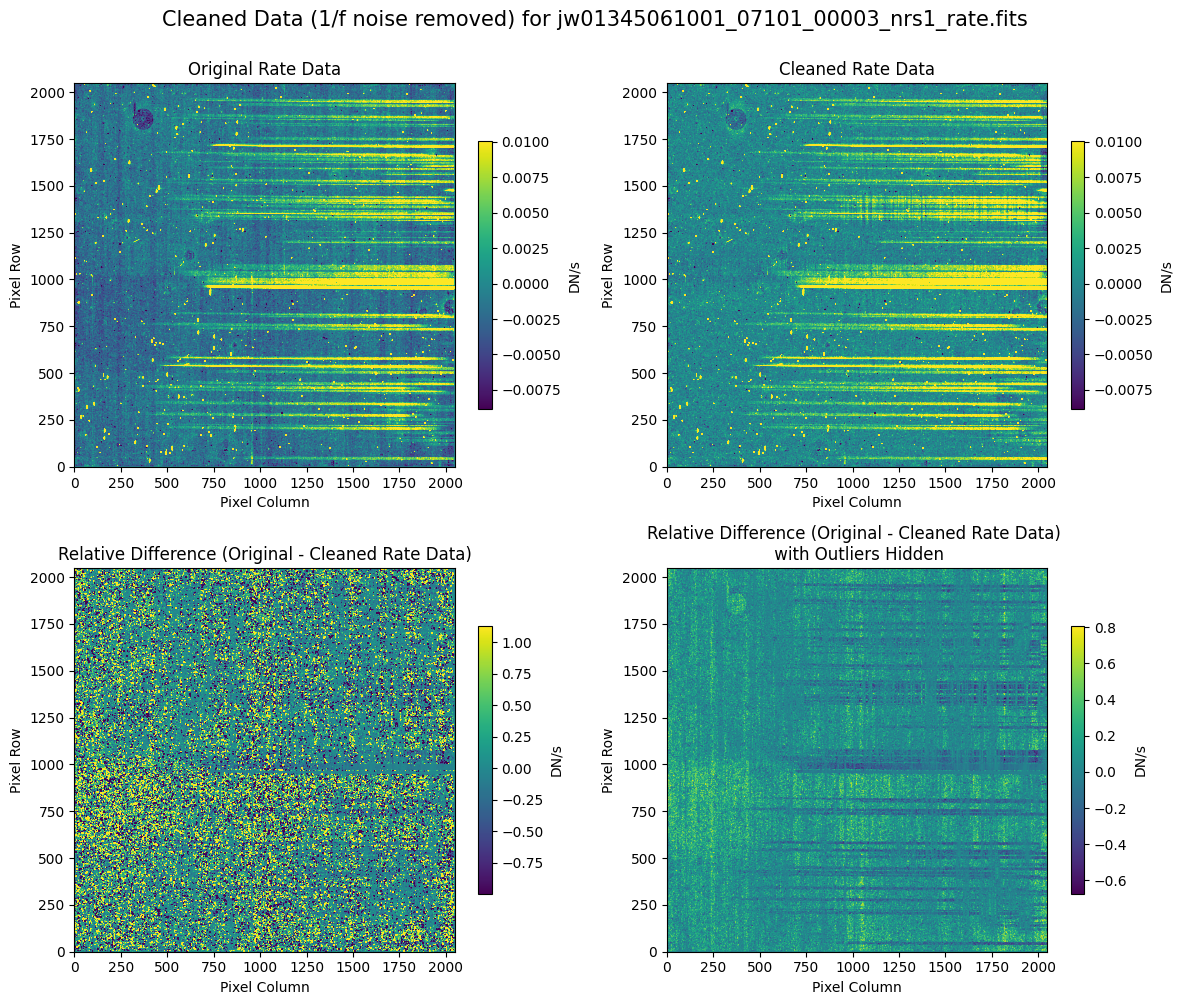
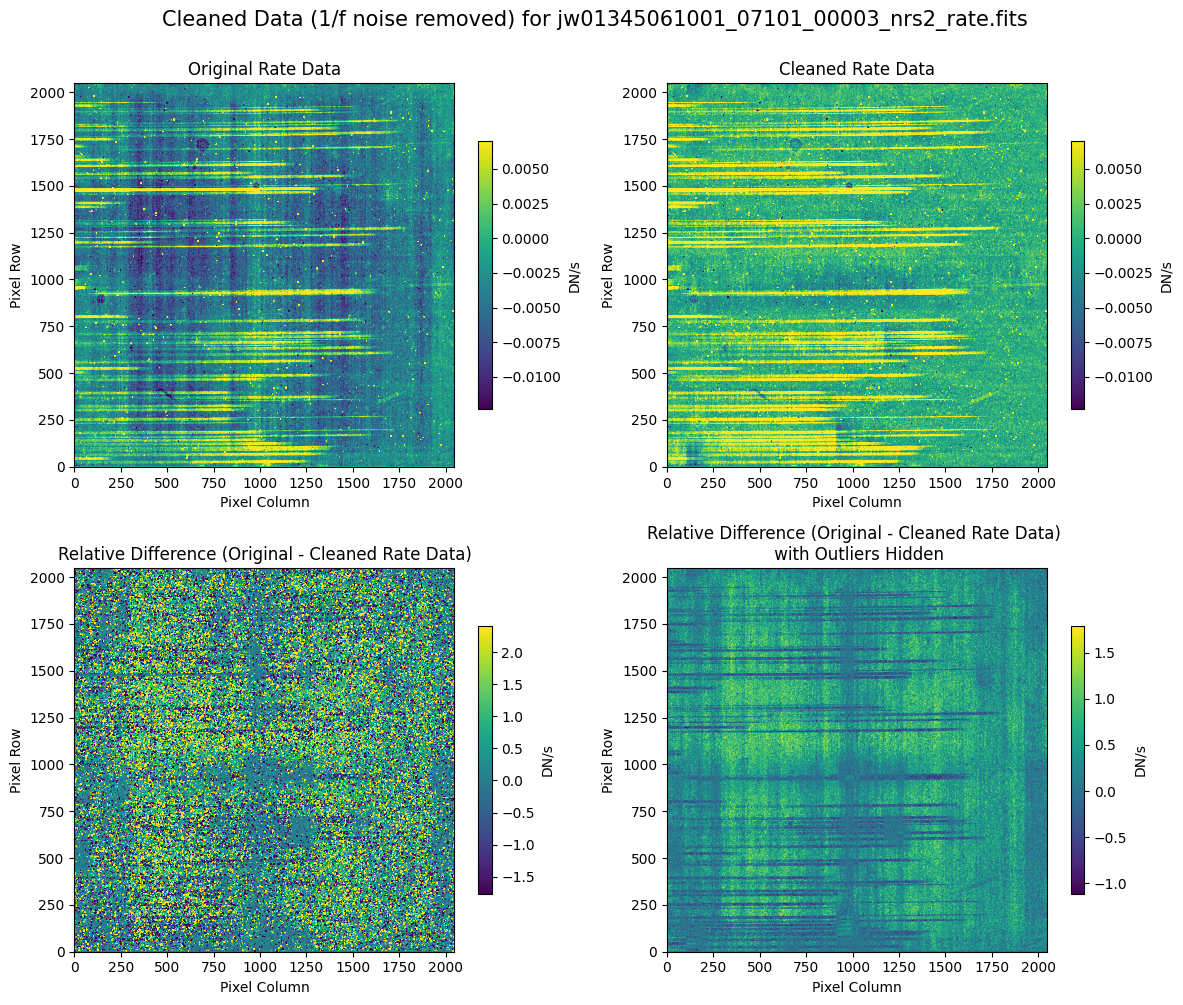
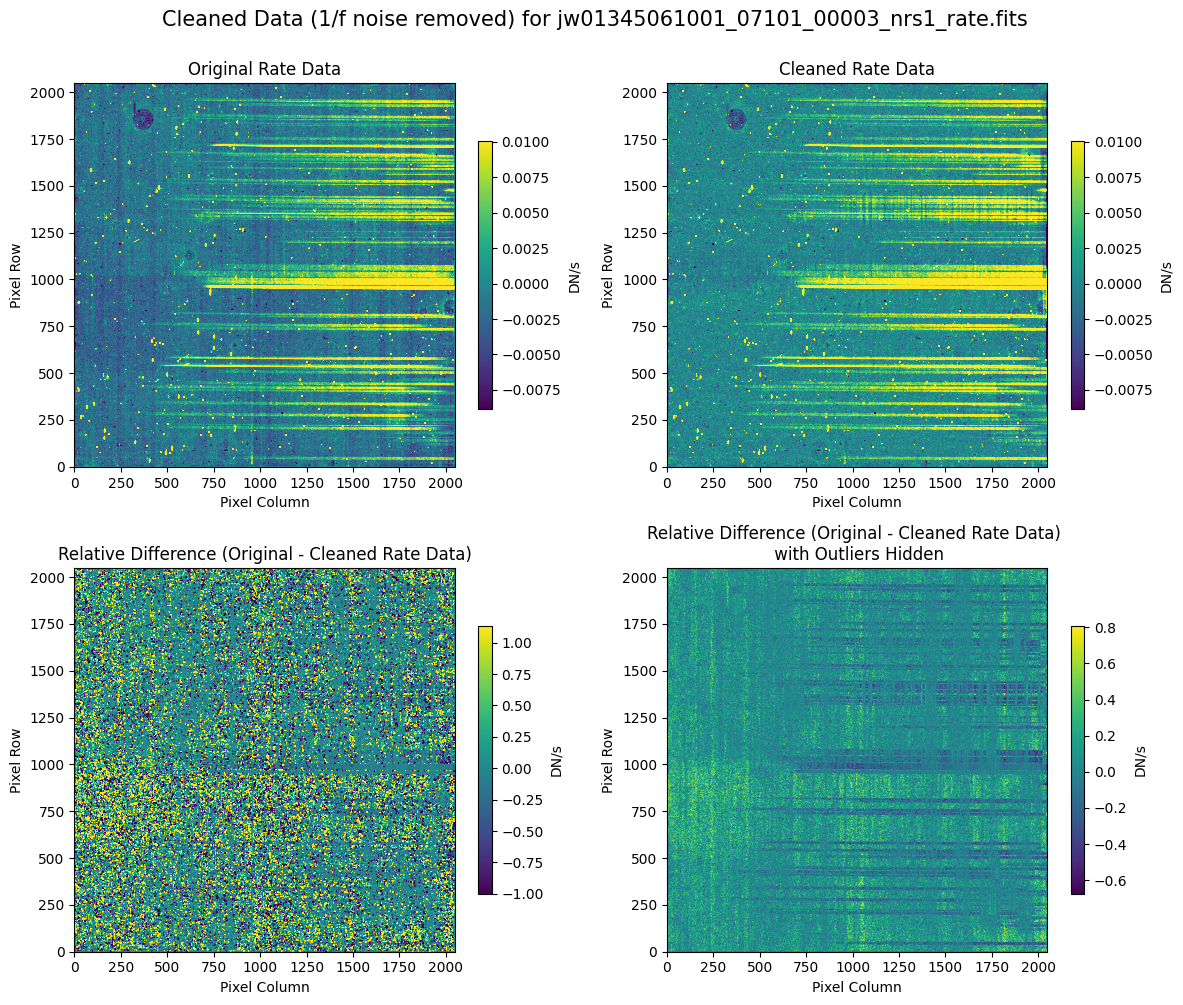
5.2 Comparing Original vs. Cleaned Data (Default Pipeline Mask) #
We can now compare the cleaned data (with the default pipeline mask) to the original rate file and verify that the 1/f noise has been reduced.
In many cases, the cleaning process introduces new artifacts to the rate file. These should be carefully examined and weighed against the benefits of noise reduction. If transient artifacts, like snowballs, are interfering with the cleaning process, it may be beneficial to manually edit the mask to remove these areas from consideration in the background fit. To do so, try varying the outlier detection threshold or editing specific pixels in the mask array directly (explored in the next few sections). Otherwise, refer to the NSClean documentation for additional suggestions on manual editing.
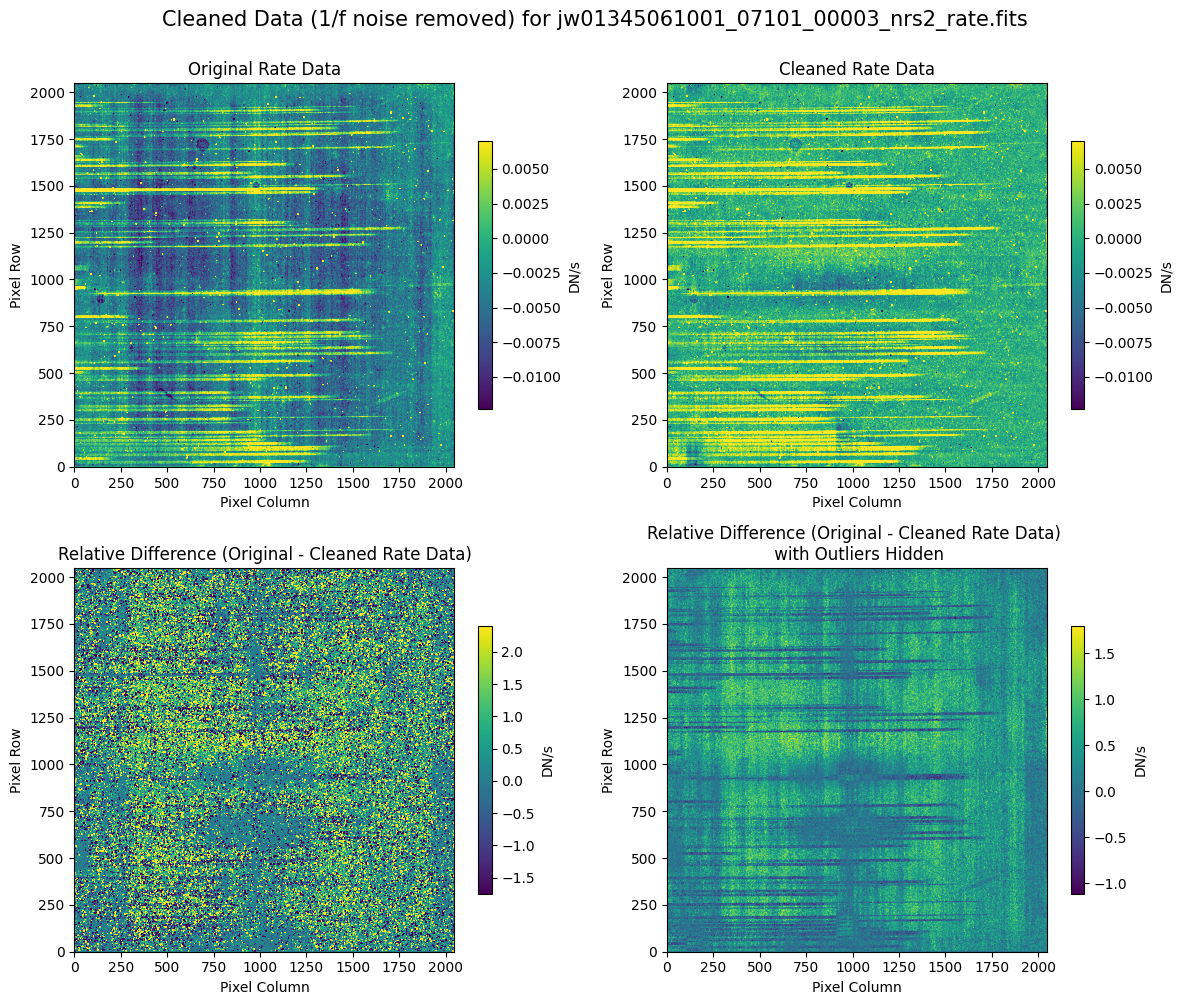
Note that in the images below, there are scattered values with large relative differences from the original rate file (shown in the relative difference image below). These are artifacts of the cleaning process.
There are also broader low-level residual background effects (shown in the relative difference image on the right, below, with scattered outliers, identified with sigma clipping, hidden by masking). These include the background patterns we are trying to remove: the 1/f noise variations in the dispersion direction and the picture frame effect at the top and bottom of the frame (for full-frame data). However, there may also be low-level artifacts introduced by over-fitting the dark data in the cleaning process.
Check both residual images carefully to understand the impact of the cleaning process on your data.
# Plot the original and cleaned data, as well as a residual map.
cleaned_default_masks = [
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs1_nsclean.fits",
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs2_nsclean.fits",
]
# Plot each associated set of rateint data and cleaned file.
for rate_file, cleaned_file in zip(rate_names, cleaned_default_masks):
plot_cleaned_data(
mast_products_dir + rate_file, cleaned_file, layout="columns", scale=9
)
# Plot heavily masked region around x=1000, y=600 NRS2
# Masking can introduce some high frequency noise in the cleaning process that
# appears as vertical striping over the spectral traces
rate_file = mast_products_dir + rate_names[1] # NRS2
original_rate_data = fits.open(rate_file)[1].data[550:651, 800:1401]
cleaned_rate_data = fits.open(cleaned_default_masks[1])[1].data[550:651, 800:1401]
# For plotting vizualization
original_rate_data[np.isnan(original_rate_data)] = 0
cleaned_rate_data[np.isnan(cleaned_rate_data)] = 0
vmin = np.nanpercentile(cleaned_rate_data, 5)
vmax = np.nanpercentile(cleaned_rate_data, 100-9)
# Original vs. cleaned data (with default mask)
fig, axs = plt.subplots(1, 2, figsize=(15, 4))
fig.colorbar(
axs[0].imshow(
original_rate_data,
cmap="viridis",
aspect=4,
origin="lower",
clim=(vmin, vmax),
),
ax=axs[0],
pad=0.05,
shrink=0.7,
label="DN/s",
)
fig.colorbar(
axs[1].imshow(
cleaned_rate_data,
cmap="viridis",
aspect=4,
origin="lower",
clim=(vmin, vmax),
),
ax=axs[1],
pad=0.05,
shrink=0.7,
label="DN/s",
)
# Set titles, tick values, xlabel, and ylabel for subplots
for ax, title in zip(axs, ["Original Rate Data (NRS2)", "Cleaned Rate Data (NRS2)"]):
ax.set(title=title, xlabel="Pixel Column", ylabel="Pixel Row")
ax.set_xticklabels([700, 800, 900, 1000, 1100, 1200, 1300, 1400])
ax.set_yticklabels([475, 500, 525, 575, 600, 625, 650])
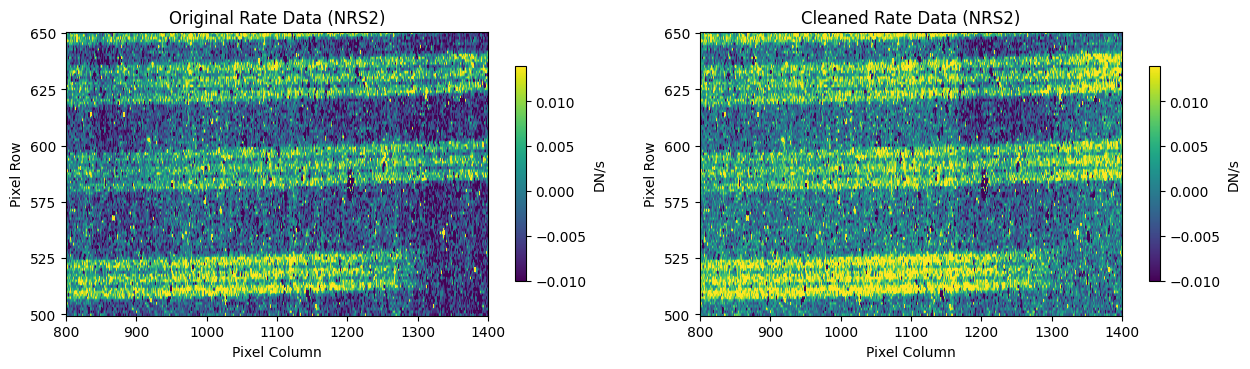
Compare the extracted spectrum from the cleaned data to the spectrum extracted from the original rate file.
# 1D extracted spectra.
x1d_nsclean_skipped = [
stage2_nsclean_skipped_dir + "jw01345061001_07101_00003_nrs1_x1d.fits",
stage2_nsclean_skipped_dir + "jw01345061001_07101_00003_nrs2_x1d.fits",
]
x1d_nsclean_default = [
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs1_x1d.fits",
stage2_nsclean_default_dir + "jw01345061001_07101_00003_nrs2_x1d.fits",
]
# Wavelength region of interest.
for original, cleaned in zip(x1d_nsclean_skipped, x1d_nsclean_default):
plot_spectra([original, cleaned], scale_percent=9)
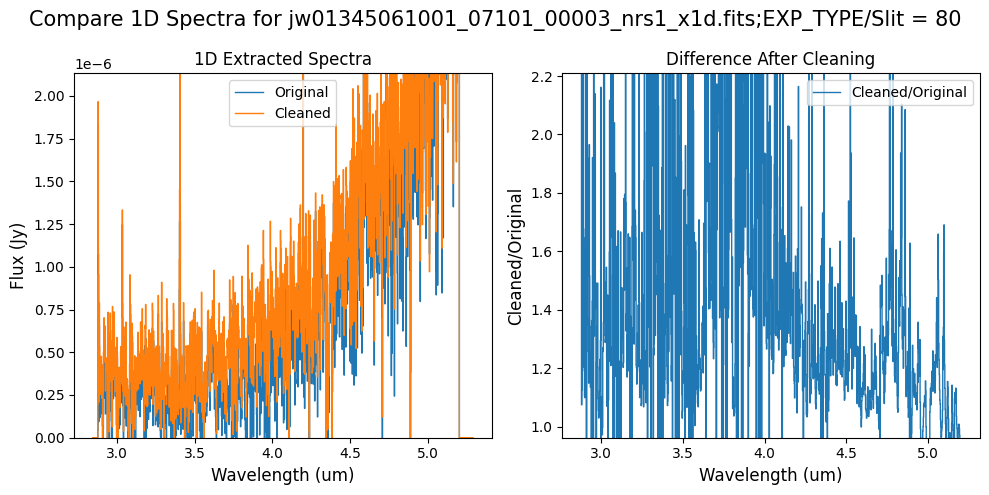
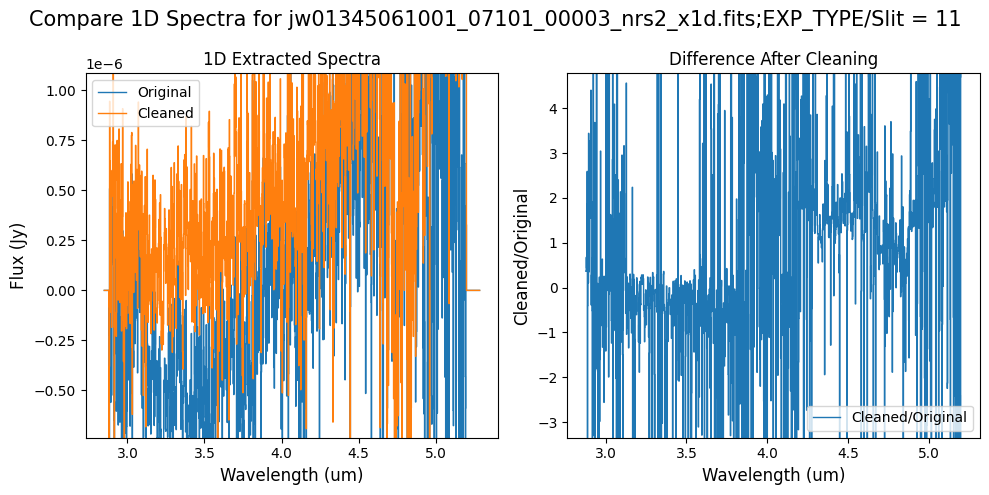
Notes:
In slit 80 on NRS1 and in slit 11 on NRS2, the overall continuum level has been slightly altered by the cleaning process.
In slit 11 on NRS2, some of the negative flux in the original spectrum has been corrected. However, excessive masking in the default mask has introduced high-frequency noise, particularly affecting slit 11 between 4.5 - 5.0 um. This results in a noticeable dip in the spectrum extracted from the cleaned data compared to the original raw data (above).
6. Clean up 1/f Noise with NSClean (Alternate Mask) #
For this data set, note that some regions of the detector are heavily masked, due to overlapping slit regions. For example, see the cleaned rate data for NRS2 above, around x=1000, y=600.
In this region, the cleaning process introduces some high frequency noise that appears as vertical striping over the spectral traces. Slit 11 is extracted from this region, and shows a noticeable dip in the spectrum extracted from the cleaned data, compared to the original rate data (above).
Also note that for MOS data, there may be several illuminated regions of the detector that are not masked by the slitlet bounding boxes. For the M gratings, zeroth-order spectra may appear on the detector, and are not easily located. For the long-pass filters, there is still some light past the red cutoff of the slitlet bounding box.
In this case, it may be beneficial to build the mask with an alternate algorithm. Here, we do not use slitlet bounding boxes and instead iteratively mask any data more than 1 sigma above the background. This leaves more dark data between the spectral traces and improves the background fit in the problematic area.
Note, however, that excessive cleaning may impact the continuum level for the spectra, if too much or too little illuminated data is included in the mask. Again, the generated mask and output spectra should be carefully examined to weigh the benefits of cleaning against the impact on the spectra.
To tune the illumination detection in this mask, try modifying the n_sigma parameter below. A higher value will identify less illumination. A lower value will identify more.
# Set up directory for running NSClean with alternate parameters.
stage2_nsclean_alternate_dir = "./stage2_nsclean_alternate/"
if not os.path.exists(stage2_nsclean_alternate_dir):
os.makedirs(
stage2_nsclean_alternate_dir
) # Create the directory if it doesn't exist.
# 1/f noise cleaned data (alternate NSClean pipeline mask).
# Estimated run time: 7 minutes.
start = tt.time()
for indx, i in enumerate(rate_names):
print(f"Processing {i}...")
if "nrs1" in i:
slit_name = "80"
else:
slit_name = "11"
Spec2Pipeline.call(
mast_products_dir + i,
save_results=True,
steps={
"nsclean": {
"skip": False,
"save_mask": True,
"save_results": True,
"n_sigma": 1,
"mask_spectral_regions": False,
},
"extract_2d": {"slit_name": slit_name},
},
output_dir=stage2_nsclean_alternate_dir,
)
print(f"Saved {i[:-9]}" + "mask.fits")
print(f"Saved {i[:-9]}" + "nsclean.fits")
print(f"Saved {i[:-9]}" + "cal.fits")
print(f"Saved {i[:-9]}" + "x1d.fits")
end = tt.time()
print("Run time: ", round(end - start, 1) / 60.0, " min")
Processing jw01345061001_07101_00003_nrs1_rate.fits...
Saved jw01345061001_07101_00003_nrs1_mask.fits
Saved jw01345061001_07101_00003_nrs1_nsclean.fits
Saved jw01345061001_07101_00003_nrs1_cal.fits
Saved jw01345061001_07101_00003_nrs1_x1d.fits
Processing jw01345061001_07101_00003_nrs2_rate.fits...
Saved jw01345061001_07101_00003_nrs2_mask.fits
Saved jw01345061001_07101_00003_nrs2_nsclean.fits
Saved jw01345061001_07101_00003_nrs2_cal.fits
Saved jw01345061001_07101_00003_nrs2_x1d.fits
Run time: 2.8133333333333335 min
6.1 Verify the Mask (Alternate Mask) #
Check the mask against the rate data to make sure it keeps only dark areas of the detector.
# Plot the rate data with masked areas blocked.
# List of on-the-fly built masks from the pipeline.
nsclean_alternate_masks = [
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs1_mask.fits",
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs2_mask.fits",
]
# Plot each associated set of rateint data and mask file.
for rate_file, mask_file in zip(rate_names, nsclean_alternate_masks):
plot_dark_data(mast_products_dir + rate_file, mask_file, layout="columns", scale=9)
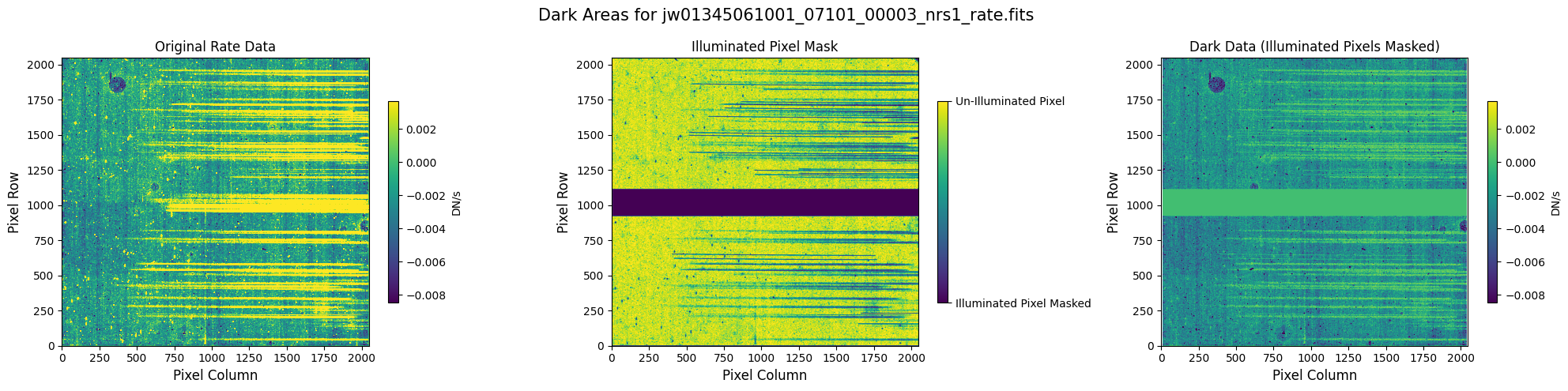
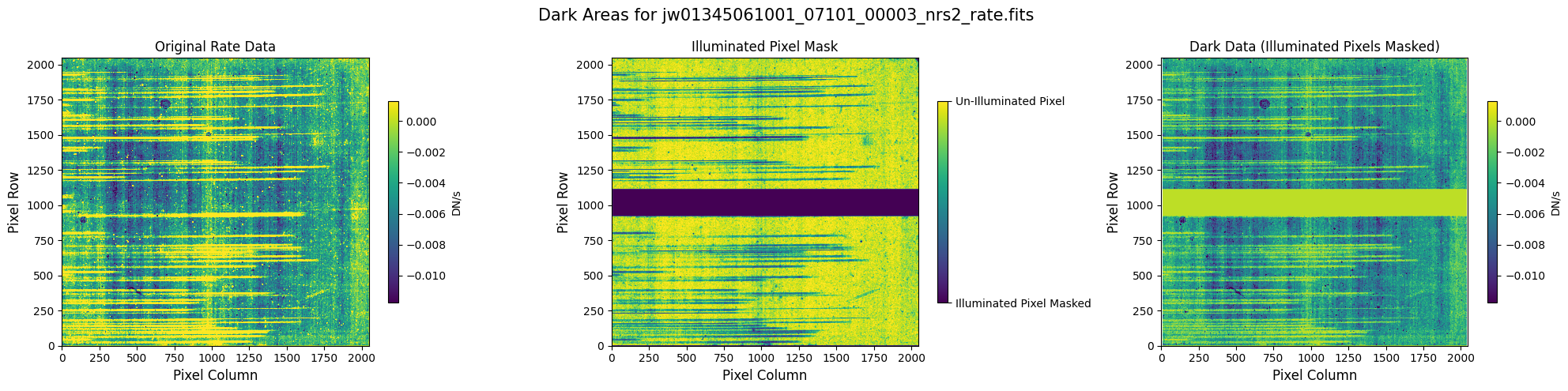
6.2 Comparing Original vs. Cleaned Data (Alternate Mask) #
# Plot the original and cleaned data, as well as a residual map.
cleaned_alternate_masks = [
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs1_nsclean.fits",
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs2_nsclean.fits",
]
# Plot each associated set of rateint data and cleaned file.
for rate_file, cleaned_file in zip(rate_names, cleaned_alternate_masks):
plot_cleaned_data(
mast_products_dir + rate_file, cleaned_file, layout="columns", scale=9
)
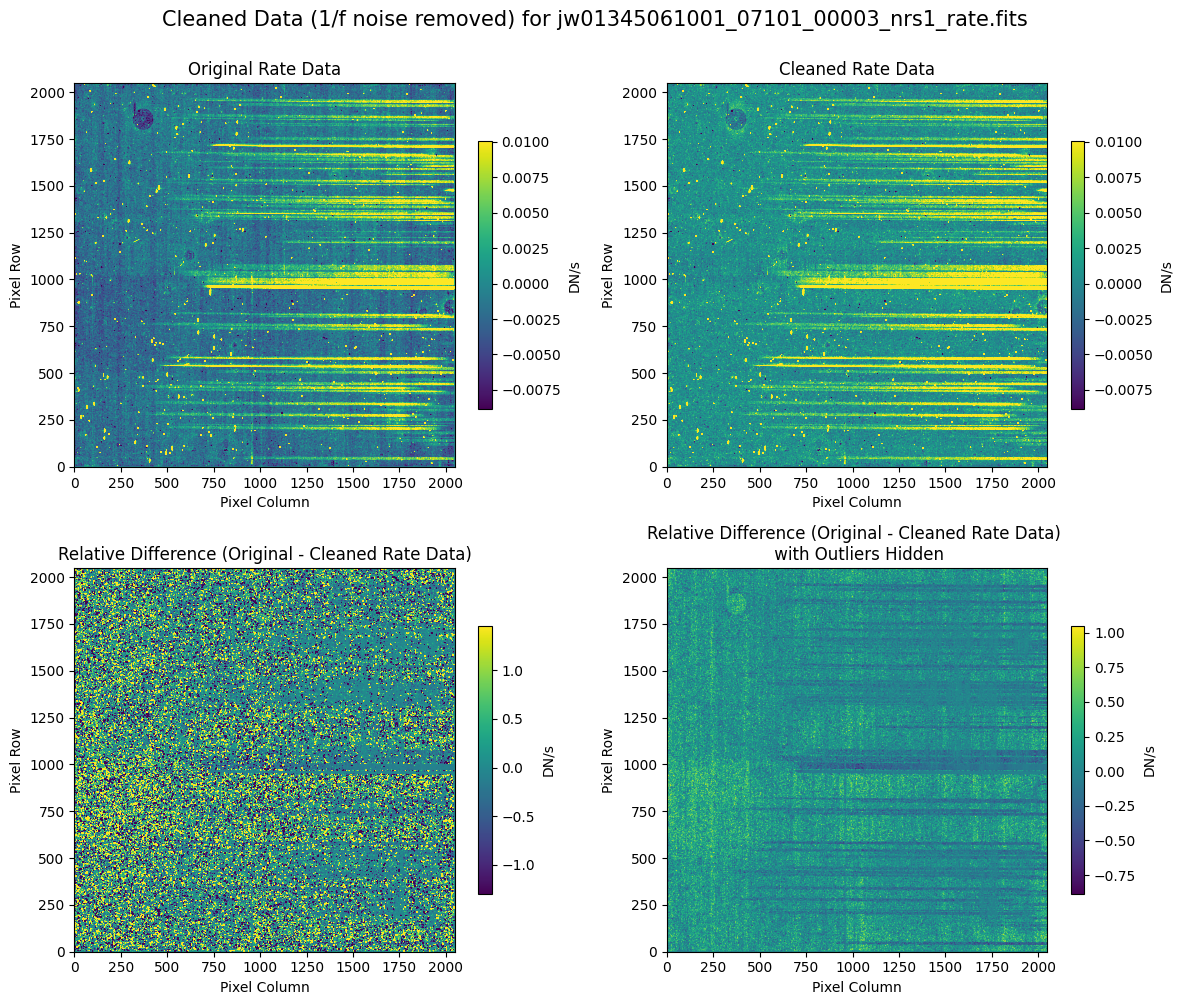
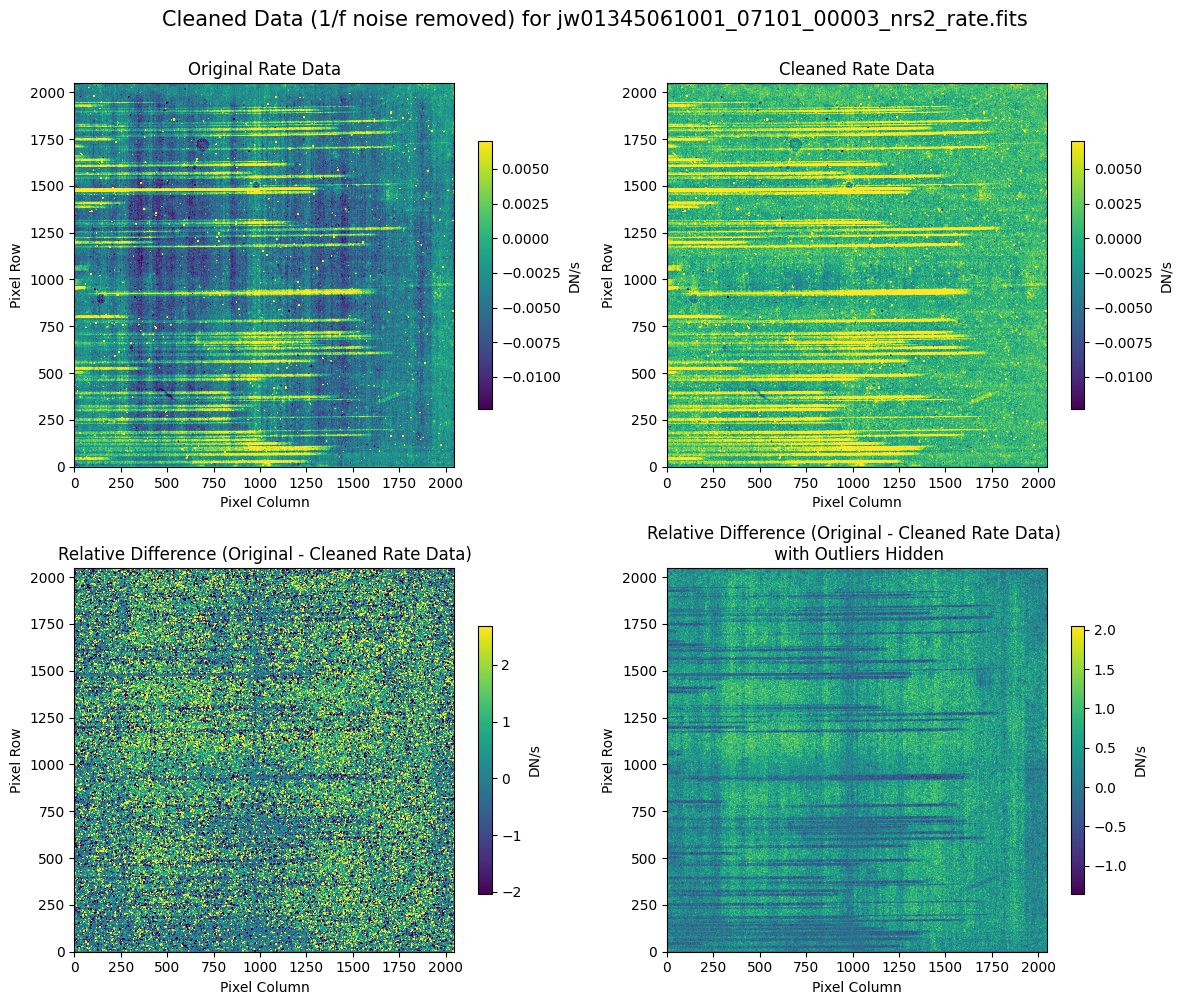
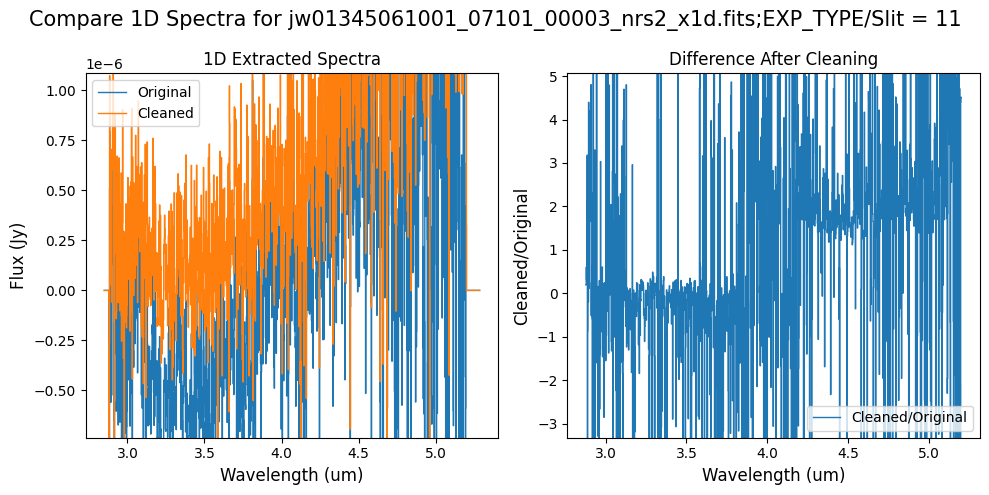
Compare the extracted spectrum from the cleaned data to the spectrum extracted from the original rate file.
x1d_nsclean_alternate = [
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs1_x1d.fits",
stage2_nsclean_alternate_dir + "jw01345061001_07101_00003_nrs2_x1d.fits",
]
for original, cleaned in zip(x1d_nsclean_skipped, x1d_nsclean_alternate):
plot_spectra([original, cleaned], scale_percent=9)
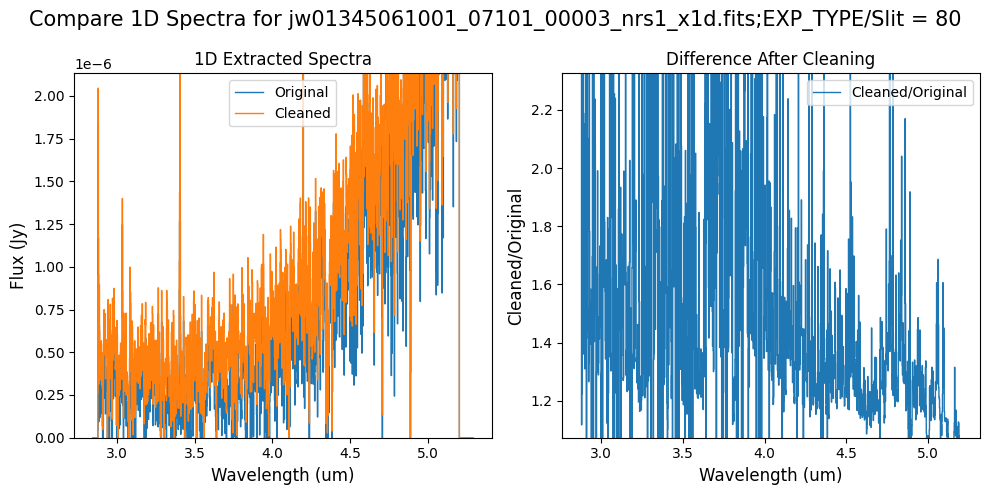
Notes:
In slit 80 on NRS1 and in slit 11 on NRS2, the overall continuum level has been slightly altered by the cleaning process (similar to the default masking).
In slit 11 on NRS2, some of the negative flux in the original spectrum has been corrected. The high-frequency noise introduced by the default masking, which affected slit 11 between 4.5 - 5.0 um (resulting in a negative dip in the spectrum), is improved with the alternate masking.
7. Clean up 1/f Noise with NSClean (Hand-Modified Mask) #
In certain scenarios, manual generation of a mask may be required. Here, we present **one** approach to manually modify the mask (editing an overly masked region in NRS2 around x=1000, y=600 to include more background; excluding some large snowballs in NRS1), starting with the default mask output from the pipeline. It is worth noting that the mask modified using this method may not necessarily outperform the two previous options.
# Set up directory for running NSClean with user-supplied mask.
stage2_nsclean_modified_dir = "./stage2_nsclean_modified/"
if not os.path.exists(stage2_nsclean_modified_dir):
# Create the directory if it doesn't exist.
os.makedirs(stage2_nsclean_modified_dir)
# Hand-modify certain mask regions.
# Specifically modifying the region in NRS2 around x=1000, y=600.
# Define the list to store paths of modified masks.
nsclean_modified_masks = []
# Iterate through the list of original masks.
for mask in nsclean_default_masks:
# New mask file name.
output_file = os.path.basename(mask)[:-5] + "_modified.fits"
# Open the FITS file.
with fits.open(mask) as hdul:
# Extract the mask data from the science extension.
mask_data = hdul["SCI"].data.copy() # Make a copy.
if "nrs2" in mask:
# Step 1: Set the default masked regions back to True.
mask_data[550:651, :1300] = True
# Step 2: Re-define masked regions by hand.
mask_data[550:575, :1300] = False # Crowded region NRS2.
mask_data[590:615, 150:1720] = False
mask_data[622:647, 100:1620] = False
else:
mask_data[50:130, 780:850] = False # Snowballs
mask_data[110:140, 920:945] = False
mask_data[820:940, 2000:2040] = False
mask_data[1650:1700, 1900:1960] = False
mask_data[1800:1900, 330:410] = False
# Update the data within the science extension.
hdul["SCI"].data = mask_data
# Save the modified FITS file
output_path = os.path.join(stage2_nsclean_modified_dir, output_file)
hdul_modified = hdul.copy() # Make a copy.
hdul_modified.writeto(output_path, overwrite=True)
nsclean_modified_masks.append(output_path)
print(f"Saved modified mask as: {output_path}")
Saved modified mask as: ./stage2_nsclean_modified/jw01345061001_07101_00003_nrs1_mask_modified.fits
Saved modified mask as: ./stage2_nsclean_modified/jw01345061001_07101_00003_nrs2_mask_modified.fits
7.1 Verify the Mask (Hand-Modified Mask) #
Check the mask against the rate data to make sure it keeps only dark areas of the detector.
# Plot the rate data with masked areas blocked.
# List of modified masks for the pipeline.
nsclean_modified_masks = [
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs1_mask_modified.fits",
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs2_mask_modified.fits",
]
# Plot each associated set of rateint data and mask file.
for rate_file, mask_file in zip(rate_names, nsclean_modified_masks):
plot_dark_data(mast_products_dir + rate_file, mask_file, layout="columns", scale=9)
# 1/f noise cleaned data (user-supplied mask).
# Estimated run time: 5 minutes.
start = tt.time()
for indx, i in enumerate(rate_names):
print(f"Processing {i}...")
if "nrs1" in i:
slit_name = "80"
else:
slit_name = "11"
Spec2Pipeline.call(
mast_products_dir + i,
save_results=True,
steps={
"nsclean": {
"skip": False,
"save_mask": True,
"save_results": True,
"user_mask": nsclean_modified_masks[indx],
},
"extract_2d": {"slit_name": slit_name},
},
output_dir=stage2_nsclean_modified_dir,
)
print(f"Saved {i[:-9]}" + "mask.fits")
print(f"Saved {i[:-9]}" + "nsclean.fits")
print(f"Saved {i[:-9]}" + "cal.fits")
print(f"Saved {i[:-9]}" + "x1d.fits")
end = tt.time()
print("Run time: ", round(end - start, 1) / 60.0, " min")
Processing jw01345061001_07101_00003_nrs1_rate.fits...
Saved jw01345061001_07101_00003_nrs1_mask.fits
Saved jw01345061001_07101_00003_nrs1_nsclean.fits
Saved jw01345061001_07101_00003_nrs1_cal.fits
Saved jw01345061001_07101_00003_nrs1_x1d.fits
Processing jw01345061001_07101_00003_nrs2_rate.fits...
Saved jw01345061001_07101_00003_nrs2_mask.fits
Saved jw01345061001_07101_00003_nrs2_nsclean.fits
Saved jw01345061001_07101_00003_nrs2_cal.fits
Saved jw01345061001_07101_00003_nrs2_x1d.fits
Run time: 2.191666666666667 min
7.2 Comparing Original vs. Cleaned Data (Hand-Modified Mask) #
# Plot the original and cleaned data, as well as a residual map.
cleaned_modified_masks = [
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs1_nsclean.fits",
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs2_nsclean.fits",
]
# Plot each associated set of rateint data and cleaned file.
for rate_file, cleaned_file in zip(rate_names, cleaned_modified_masks):
plot_cleaned_data(
mast_products_dir + rate_file, cleaned_file, layout="columns", scale=9
)
Compare the extracted spectrum from the cleaned data to the spectrum extracted from the original rate file.
x1d_nsclean_modified = [
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs1_x1d.fits",
stage2_nsclean_modified_dir + "jw01345061001_07101_00003_nrs2_x1d.fits",
]
# Wavelength Region of interest.
for original, cleaned in zip(x1d_nsclean_skipped, x1d_nsclean_modified):
plot_spectra([original, cleaned], scale_percent=9)
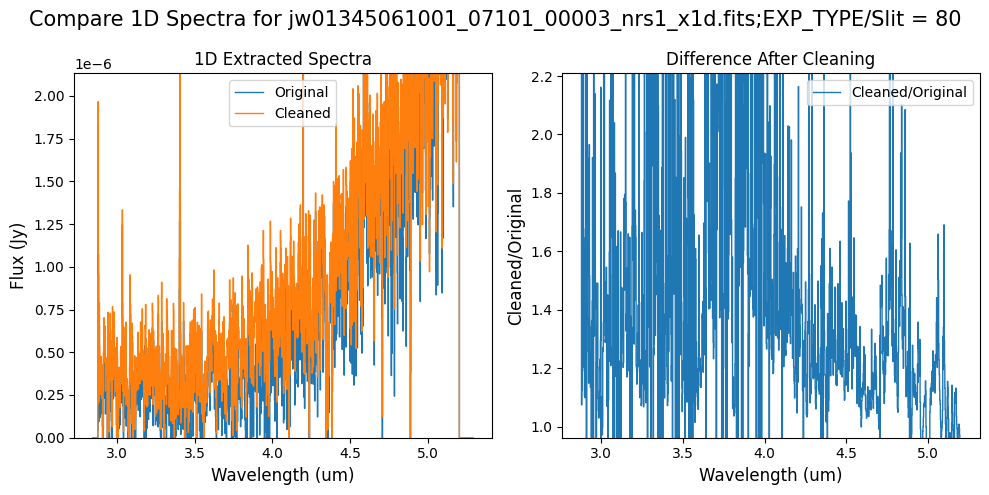
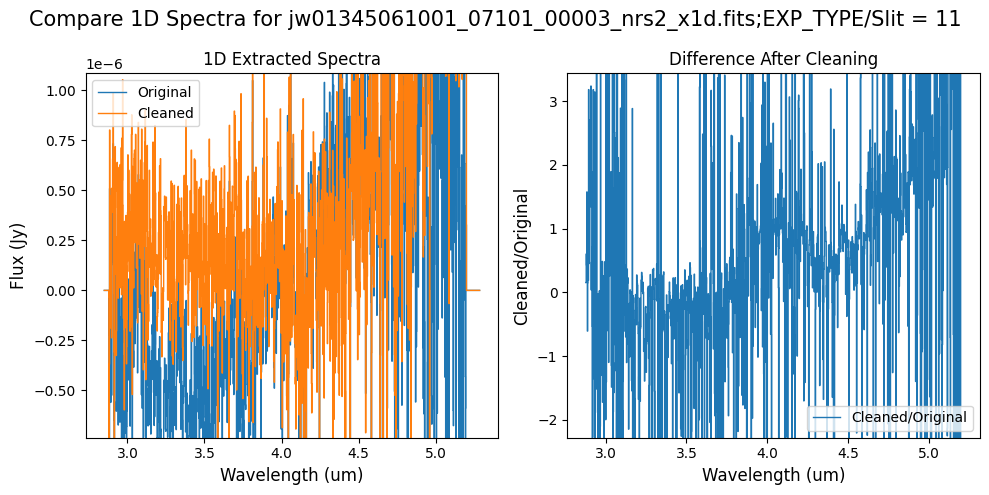
Notes:
In slit 80 on NRS1, the overall continuum level has been slightly altered by the cleaning process (similar to the default masking and alternate masking).
In slit 11 on NRS2, the flux between 4-4.55 um has decreased due to the cleaning process with the hand-modified mask. Some of the negative flux in the original spectrum (at shorter and longer wavelengths) has been corrected.
8. Conclusion #
The final plots below show the countrate images and the resulting 1D extracted spectra side-by-side to compare the different cleaning methods: the original (no NSClean applied), the cleaned countrate image (with the default pipeline mask), the cleaned countrate image (with an alternate pipeline mask), and finally, the cleaned countrate image (with the hand-modified mask).
Please note that the results presented in this notebook may vary for different datasets (e.g., targets of different brightness, spatial extent, etc.). Users are encouraged to explore NSClean using different masking methods to determine the optimal results.
The output from the cleaning algorithm is now ready for further processing. The (_cal.fits) files produced by the above Spec2Pipeline run may be used as input to the Spec3Pipeline, for generating final combined spectra.
# Not cleaned vs. cleaned (default mask) vs. cleaned (alternate mask) rate data
original_rate_data = [
fits.open(mast_products_dir + rate_name)[1].data for rate_name in rate_names
]
cleaned_rate_default_data = [
fits.open(cleaned_default_mask)[1].data
for cleaned_default_mask in cleaned_default_masks
]
cleaned_rate_alternate_data = [
fits.open(cleaned_alternate_mask)[1].data
for cleaned_alternate_mask in cleaned_alternate_masks
]
cleaned_rate_modified_data = [
fits.open(cleaned_modified_mask)[1].data
for cleaned_modified_mask in cleaned_modified_masks
]
# For plotting visualization
for data_list in [
original_rate_data,
cleaned_rate_default_data,
cleaned_rate_alternate_data,
cleaned_rate_modified_data,
]:
for data in data_list:
data[np.isnan(data)] = 0
# Original vs. cleaned data (with default mask)
fig, axs = plt.subplots(2, 4, figsize=(25, 12))
# Set y-axis titles and plot the data
titles = [
"Original Rate Data",
"Cleaned Rate Data (Default Mask)",
"Cleaned Rate Data (Alternate Mask)",
"Cleaned Rate Data (Hand-Modified Mask)",
]
for i, (data_list, title) in enumerate(
zip(
[
original_rate_data,
cleaned_rate_default_data,
cleaned_rate_alternate_data,
cleaned_rate_modified_data,
],
titles,
)
):
for j, data in enumerate(data_list):
ax = axs[j, i]
ax.set_title(f'{title} \n {"NRS1" if j == 0 else "NRS2"}', fontsize=12)
im = ax.imshow(data, origin="lower", clim=(-1e-2, 1e-2))
fig.colorbar(im, ax=ax, pad=0.05, shrink=0.7, label="DN/s")
ax.set_xlabel("Pixel Column", fontsize=10)
ax.set_ylabel("Pixel Row", fontsize=10)
plt.tight_layout()
plt.show()
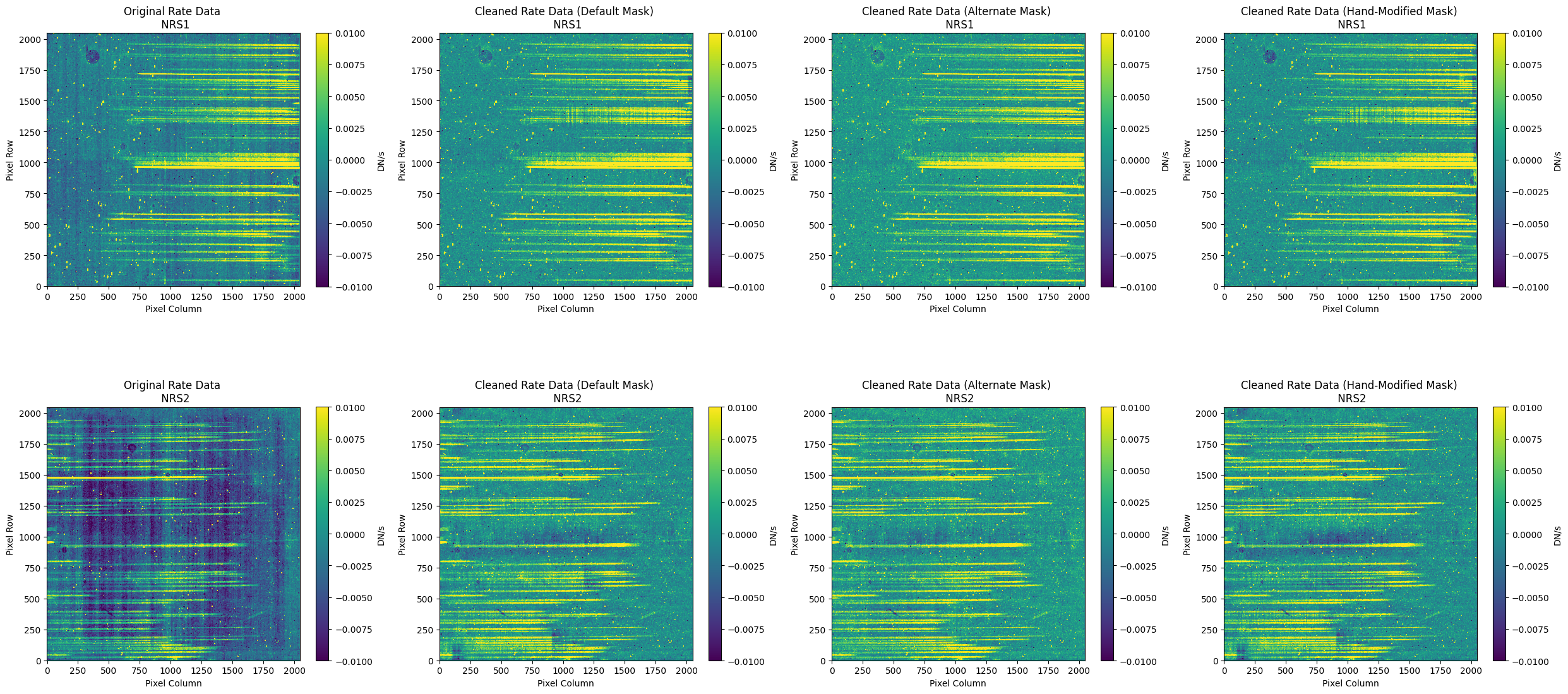
# Final Comparison
plot_spectra(
[
x1d_nsclean_skipped[0],
x1d_nsclean_default[0],
x1d_nsclean_alternate[0],
x1d_nsclean_modified[0],
],
scale_percent=4,
)
plot_spectra(
[
x1d_nsclean_skipped[1],
x1d_nsclean_default[1],
x1d_nsclean_alternate[1],
x1d_nsclean_modified[1],
],
scale_percent=4,
)
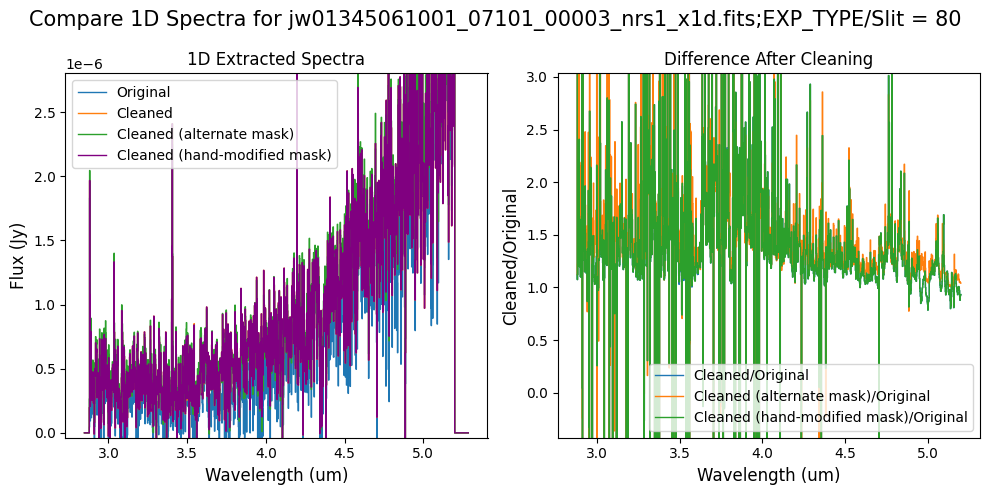
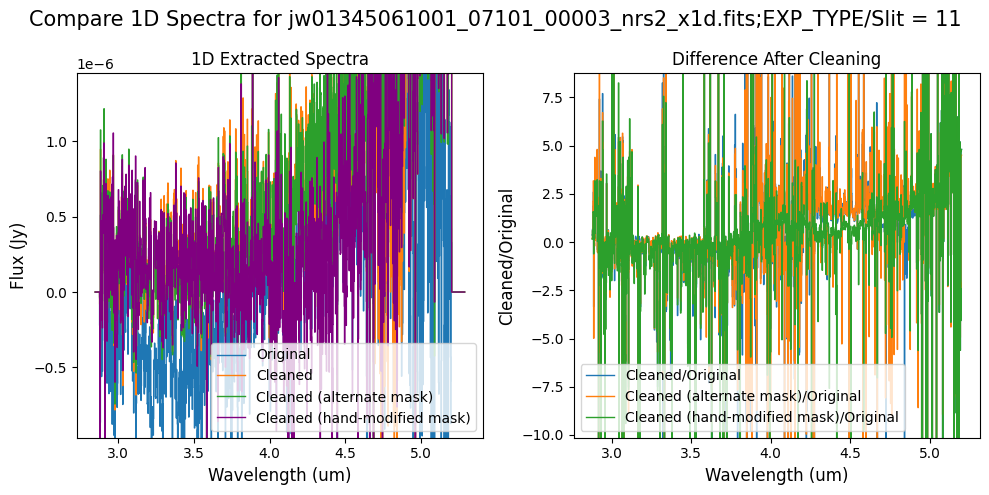
Final Notes:
The high-frequency noise in NRS2 introduced by the default masking, which was affecting the 1D extracted spectrum for slit 11 around 4.55um, no longer appears when using the alternate mask (slight improvement with the hand-modified mask), significantly improving the spectrum for slit 11.
Negative flux in slit 11 (at shorter and longer wavelengths) has been corrected with either of the masks. However, the hand-modified mask appears to decrease the continuum level for the spectrum of slit 11 between 4-4.55um. In this case, the alternate masking (clip-based) algorithm is preferable to blocking the entire science region for each MSA shutter.
About the Notebook #
Authors: Melanie Clarke, Kayli Glidic; NIRSpec Instrument Team
Updated On: Feburary 29, 2024.
