NIRSpec IFU Optimal Point Source Extraction#
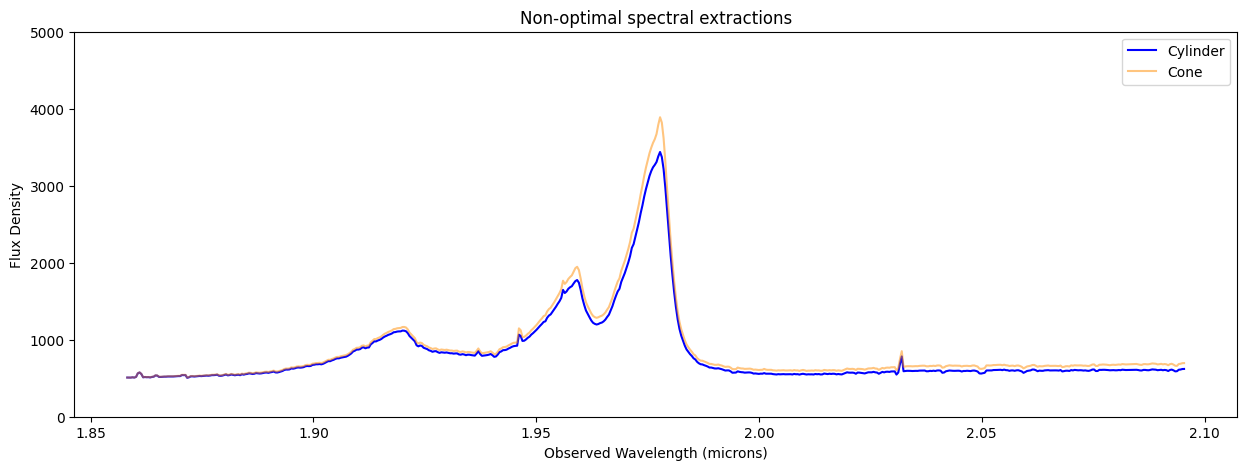
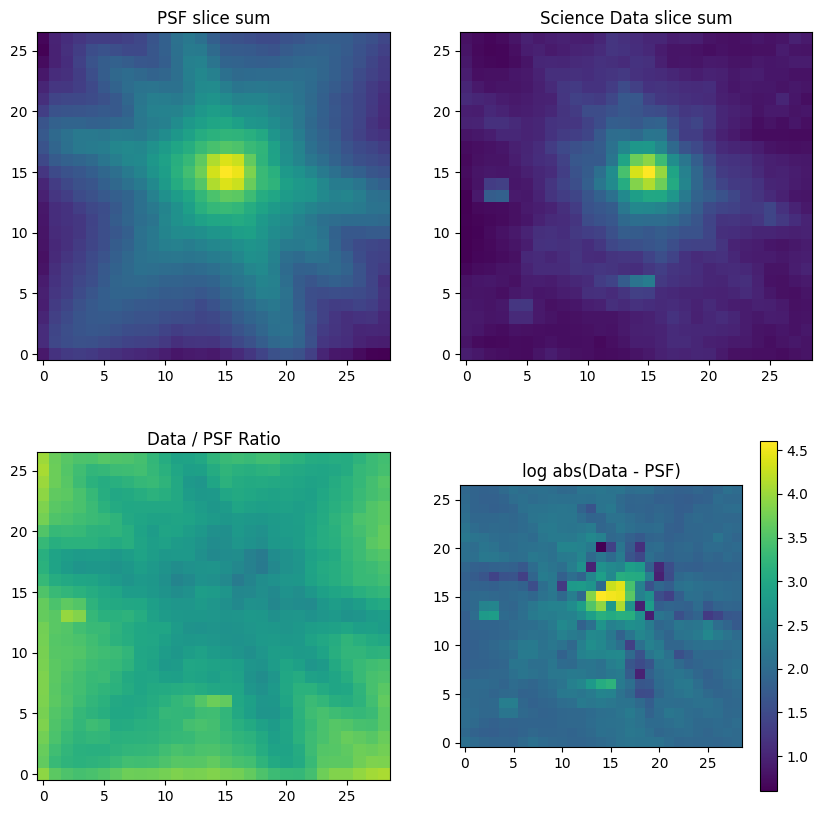
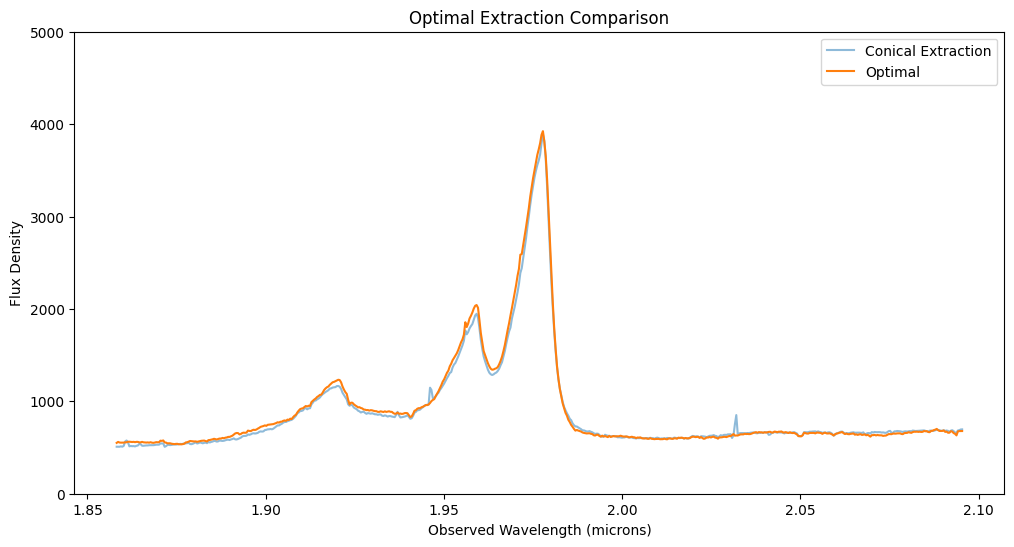
This notebook illustrates various extraction methods for a point source in JWST NIRSpec IFU data, utilizing the Q3D (PID 1335) observation of quasar SDSS J165202.64+172852.3. The extraction techniques include subset extraction with Cubeviz, simple sum over spaxels, cylindrical aperture, conical aperture photometry, and optimal point source extraction using a WebbPSF model PSF.
Use case: optimal spectral extraction; method by Horne (1986). Data: JWST NIRSpec IFU data; point sources. Tools: jwst, webbpsf, matplotlib, scipy, custom functions. Cross-intrument: any spectrograph. Documentation: This notebook is part of a STScI’s larger post-pipeline Data Analysis Tools Ecosystem.
Table of Contents#
1. Imports #
numpy for array math
scipy for ndimage shift
specutils for Spectrum1D data model
jdaviz : for data visualization
photutils to define circular apertures
astropy.io for reading and writing FITS cubes and images
astropy.wcs, units, coordinates for defining and reading WCS
astropy.stats for sigma_clipping
astropy.utils for downloading files from URLs
matplotlib for plotting spectra and images
os for file management
astroquery.mast to download the data
import numpy as np
import scipy
from specutils import Spectrum1D
import jdaviz
from jdaviz import Cubeviz, Specviz
print("jdaviz Version={}".format(jdaviz.__version__))
from photutils.aperture import CircularAperture, aperture_photometry
from astropy.io import fits
from astropy import wcs
from astropy import units as u
import os
from astroquery.mast import Observations
jdaviz Version=4.2.2
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
2. Read in NIRSpec IFU Cube #
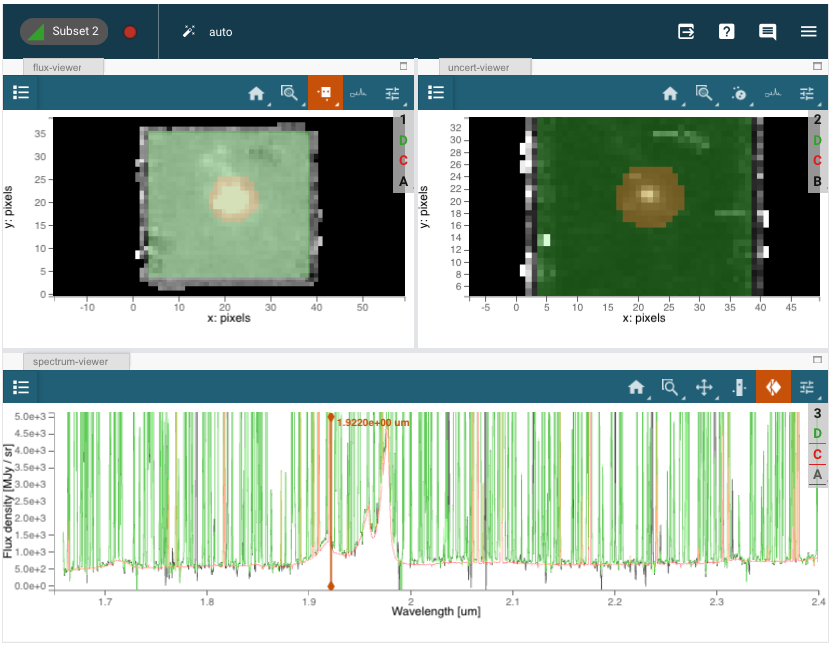
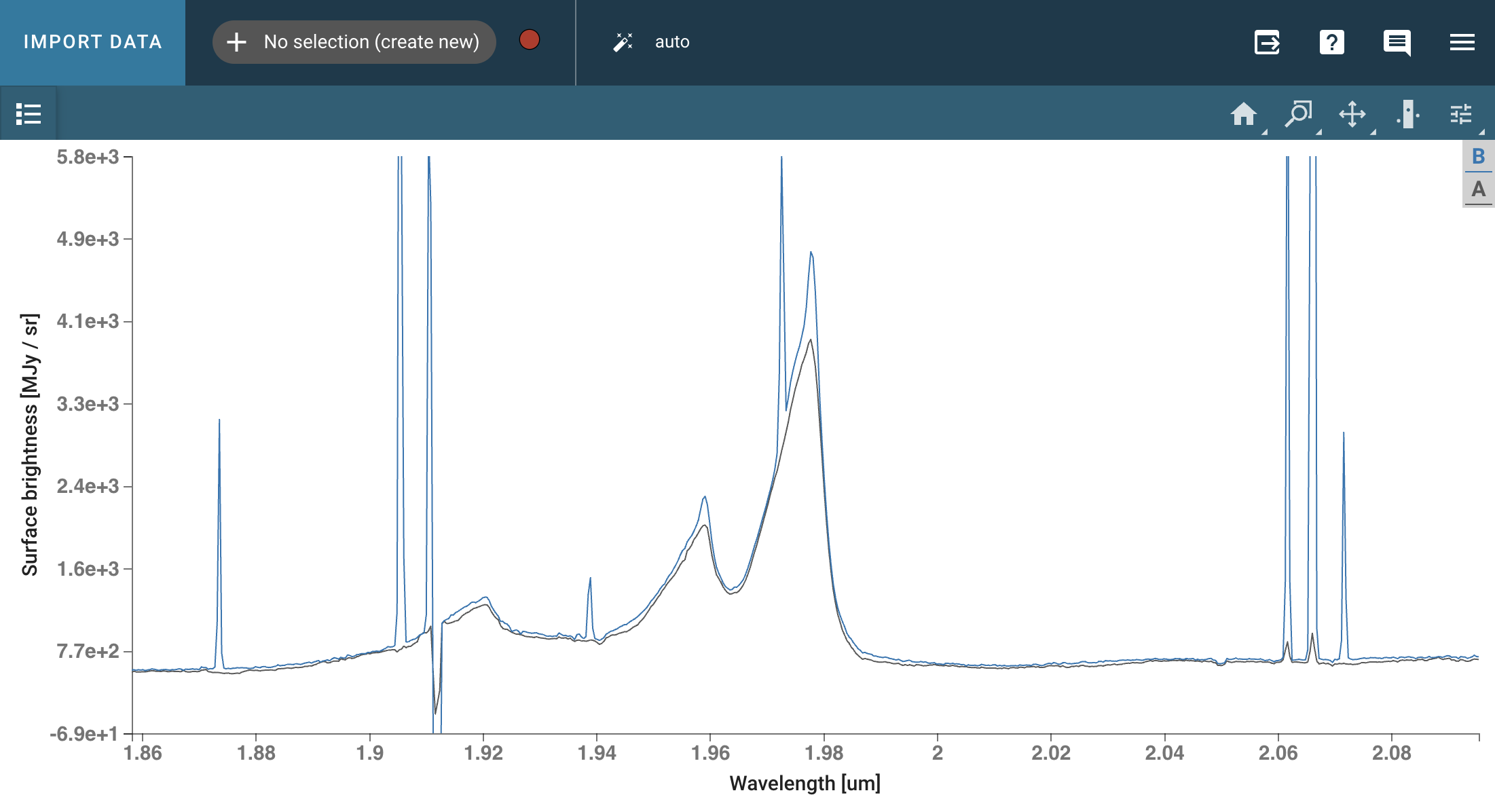
The NIRSpec IFU observation of quasar SDSS J1652+1728 (redshift z=1.9) was taken using the G235H grating with F170LP filter, covering 1.66-3.17 microns at a spectral resolution of R~2700. The IFU spaxels are 0.1” on a side. The level-3 pipeline_processed datacube (s3d.fits, which combines all dithered exposures) is retrieved from MAST in the next notebook cell below.
# Download the data file
uri = "mast:jwst/product/jw01335-o008_t007_nirspec_g235h-f170lp_s3d.fits"
result = Observations.download_file(
uri, base_url="https://mast.stsci.edu/api/v0.1/Download/file"
)
if result[0] == "ERROR":
raise RuntimeError("Error retrieving file: " + result[1])
# Construct the local filepath
filename = os.path.join(os.path.abspath("."), uri.rsplit("/", 1)[-1])
# Optionally Replace MAST data with custom reprocessed data in the current directory
# filename="./jw01335-o008_t007_nirspec_g235h-f170lp_s3d.fits"
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:jwst/product/jw01335-o008_t007_nirspec_g235h-f170lp_s3d.fits to jw01335-o008_t007_nirspec_g235h-f170lp_s3d.fits ...
[Done]
# Open and inspect the file and WCS
with fits.open(filename, memmap=False) as hdulist:
sci = hdulist["SCI"].data
err = hdulist["ERR"].data
w = wcs.WCS(hdulist[1].header)
hdr = hdulist[1].header
print(w)
# Window the wavelength range to focus on Hbeta-[OIII]
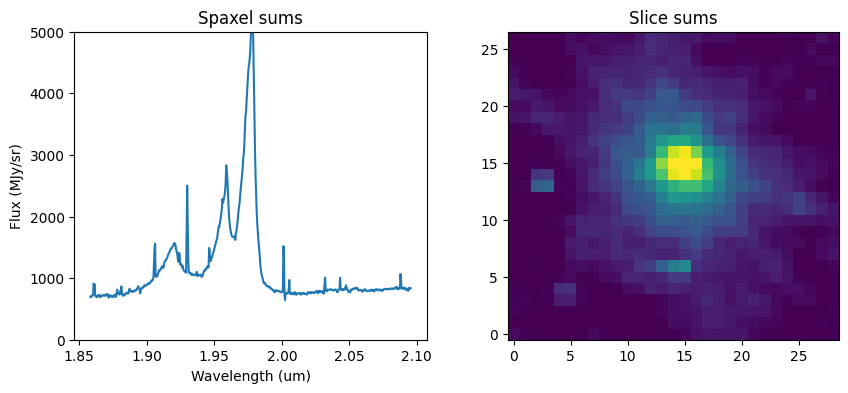
spec1d = Spectrum1D.read(filename)
slice_range = range(500, 1100, 1)
wavelength = np.array(spec1d.spectral_axis.value)[slice_range[0]: slice_range[-1] + 1]
# List of cube slices for aperture photometry
sci_data = []
sci_var = []
for idx in slice_range:
sci_data.append(sci[idx, :, :])
sci_var.append(err[idx, :, :])
data = np.nan_to_num(np.array(sci_data))
var = np.array(sci_var)
print("\nTrimmed data shape:", data.shape)
WCS Keywords
Number of WCS axes: 3
CTYPE : 'RA---TAN' 'DEC--TAN' 'WAVE'
CRVAL : -106.98898425200001 17.481222214 1.6601979666156693e-06
CRPIX : 22.0 20.0 1.0
PC1_1 PC1_2 PC1_3 : -1.0 0.0 0.0
PC2_1 PC2_2 PC2_3 : 0.0 1.0 0.0
PC3_1 PC3_2 PC3_3 : 0.0 0.0 1.0
CDELT : 2.77777781916989e-05 2.77777781916989e-05 3.95999988541007e-10
NAXIS : 43 39 3814
Trimmed data shape: (600, 39, 43)
cubeviz = Cubeviz()
cubeviz.load_data(filename)
cubeviz.show()
# Set spectrum display limits
cubeviz.specviz.x_limits(1.65 * u.um, 2.4 * u.um)
cubeviz.specviz.y_limits(0.0, 5.0e3)
# Select slice to visualize
cubeviz.select_slice(714)
 Use viewers['spectrum-viewer'].set_limits instead. [warnings]
WARNING:astroquery:AstropyDeprecationWarning: The x_limits function is deprecated and may be removed in a future version.
Use viewers['spectrum-viewer'].set_limits instead.
Use viewers['spectrum-viewer'].set_limits instead. [warnings]
WARNING:astroquery:AstropyDeprecationWarning: The x_limits function is deprecated and may be removed in a future version.
Use viewers['spectrum-viewer'].set_limits instead.
WARNING: AstropyDeprecationWarning: The y_limits function is deprecated and may be removed in a future version.
Use viewers['spectrum-viewer'].set_limits instead. [warnings]
WARNING:astroquery:AstropyDeprecationWarning: The y_limits function is deprecated and may be removed in a future version.
Use viewers['spectrum-viewer'].set_limits instead.
WARNING: AstropyDeprecationWarning: The select_slice function is deprecated and may be removed in a future version.
Use plugins['Slice'].slice instead. [warnings]
WARNING:astroquery:AstropyDeprecationWarning: The select_slice function is deprecated and may be removed in a future version.
Use plugins['Slice'].slice instead.