WFSS Spectra - Part 2: Cross Correlation to Determine Redshift#
Use case: optimal extraction of grism spectra; redshift measurement; emission-line maps. Simplified version of JDox Science Use Case # 33.
Data: JWST simulated NIRISS images from MIRAGE, run through the JWST calibration pipeline; galaxy cluster.
Tools: specutils, astropy, pandas, emcee, lmfit, corner, h5py.
Cross-intrument: NIRSpec
Documentation: This notebook is part of a STScI’s larger post-pipeline Data Analysis Tools Ecosystem.
Introduction#
This notebook is 3 of 4 in a set focusing on NIRISS WFSS data: 1. 1D optimal extraction since the JWST pipeline only provides a box extraction. Optimal extraction improves S/N of spectra for faint sources. 2. Combine and normalize 1D spectra. 3. Cross correlate galaxy with template to get redshift. 4. Spatially resolved emission line map.
This notebook derives the redshift of a galaxy with multiple emission lines by using specutils template correlation. The notebook begins with optimally extracted 1D spectra from notebook #2.
Note: Spectra without emission lines (e.g., ID 00003 in the previous notebook) may fail with the function here.
%matplotlib inline
import os
import numpy as np
import h5py
from astropy.io import fits
from astropy.table import QTable
import astropy.units as u
from astropy.nddata import StdDevUncertainty
from astropy.modeling.polynomial import Chebyshev1D
from astropy import constants as const
import astropy
print('astropy', astropy.__version__)
astropy 7.2.0
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['savefig.dpi'] = 80
mpl.rcParams['figure.dpi'] = 80
import specutils
from specutils.fitting import continuum
from specutils import Spectrum, SpectralRegion
from specutils.analysis import correlation
print("Specutils: ", specutils.__version__)
Specutils: 2.2.0
0. Download notebook 01 products#
These can be also obtained by running the notebooks.
if not os.path.exists('./output'):
import zipfile
import urllib.request
boxlink = 'https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/NIRISS_lensing_cluster/output.zip'
boxfile = './output.zip'
urllib.request.urlretrieve(boxlink, boxfile)
zf = zipfile.ZipFile(boxfile, 'r')
list_names = zf.namelist()
for item in list_names:
zf.extract(member=item) # Using extract instead of extractall because it is safer when files have absolute (/) or relative (..) paths
else:
print('Already exists')
Already exists
1.Open optimally extracted 1D spectrum text file;#
These are optimally extracted, normalized spectra from notebooks 01a & 01b.
DIR_OUT = './output/'
filt = 'f200w'
grism = 'G150C'
# grism = 'G150R'
id = '00004'
file_1d = f'{DIR_OUT}l3_nis_{filt}_{grism}_s{id}_combine_1d_opt.fits'
fd = fits.open(file_1d)[1].data
hd = fits.open(file_1d)[1].header
fd
FITS_rec([(1.71986127, 42.38365482, 1.51015508),
(1.72453201, 38.56917716, 0.97178267),
(1.72920275, 35.93947653, 0.62354777),
(1.73387337, 23.77350774, 0.31776521),
(1.73854411, 22.91664629, 0.21997059),
(1.74321485, 18.82889931, 0.14420642),
(1.74788558, 16.24401145, 0.10209115),
(1.7525562 , 12.60893081, 0.0719594 ),
(1.75722694, 10.30048011, 0.05026028),
(1.76189768, 10.56057566, 0.04827347),
(1.76656842, 10.80664315, 0.04615251),
(1.77123904, 10.96807816, 0.04635962),
(1.77590978, 10.84610055, 0.04520876),
(1.78058052, 11.14885447, 0.04861023),
(1.78525114, 11.40215898, 0.05138483),
(1.78992188, 11.48868052, 0.04729241),
(1.79459262, 11.58795191, 0.04634937),
(1.79926336, 11.73206527, 0.04663301),
(1.80393398, 11.97186637, 0.04839924),
(1.80860472, 11.9611019 , 0.04914871),
(1.81327546, 11.77717771, 0.04949575),
(1.81794608, 11.6472086 , 0.04936947),
(1.82261682, 11.56556774, 0.04957357),
(1.82728755, 11.71344915, 0.04822698),
(1.83195829, 11.65288706, 0.04838117),
(1.83662891, 11.73176765, 0.04977119),
(1.84129965, 11.98642048, 0.04941678),
(1.84597039, 12.14407666, 0.04846335),
(1.85064113, 12.2294789 , 0.04976337),
(1.85531175, 12.58098372, 0.05318308),
(1.85998249, 12.62738604, 0.05218814),
(1.86465323, 12.40292156, 0.05172025),
(1.86932385, 12.20461574, 0.05425321),
(1.87399459, 12.41531018, 0.05062367),
(1.87866533, 12.49310545, 0.0504054 ),
(1.88333607, 12.47879134, 0.05008047),
(1.88800669, 12.48052731, 0.05093631),
(1.89267743, 12.44949336, 0.0523712 ),
(1.89734817, 12.46076927, 0.0512366 ),
(1.90201879, 12.50730277, 0.05058337),
(1.90668952, 12.43958853, 0.05015841),
(1.91136026, 12.42111697, 0.05012851),
(1.916031 , 12.46504231, 0.05106978),
(1.92070162, 12.43572865, 0.05177651),
(1.92537236, 12.50323568, 0.0518209 ),
(1.9300431 , 12.56200541, 0.05140919),
(1.93471372, 12.524999 , 0.05147548),
(1.93938446, 12.59353438, 0.0513625 ),
(1.9440552 , 12.66183485, 0.05211391),
(1.94872594, 12.62118241, 0.05276501),
(1.95339656, 12.35676077, 0.05585218),
(1.9580673 , 12.31514282, 0.05852116),
(1.96273804, 12.6059867 , 0.0581315 ),
(1.96740878, 12.7973866 , 0.05491294),
(1.9720794 , 13.13971683, 0.05460986),
(1.97675014, 13.4866589 , 0.05613727),
(1.98142087, 13.52707449, 0.0561673 ),
(1.98609149, 13.5420637 , 0.05623735),
(1.99076223, 13.79659428, 0.05655741),
(1.99543297, 14.08201932, 0.05897219),
(2.00010371, 14.67188355, 0.05941912),
(2.00477433, 15.88794186, 0.06221541),
(2.00944495, 17.92468302, 0.06824417),
(2.01411581, 21.49804726, 0.0774888 ),
(2.01878643, 28.19607868, 0.08944139),
(2.02345729, 39.44270001, 0.111863 ),
(2.02812791, 54.27856681, 0.14055919),
(2.03279853, 65.73083879, 0.16328814),
(2.03746939, 65.81027175, 0.16305239),
(2.04214001, 54.96117411, 0.14097168),
(2.04681063, 40.50272461, 0.11793418),
(2.05148149, 29.101485 , 0.09050252),
(2.05615211, 22.31114997, 0.07628278),
(2.06082273, 18.71178258, 0.06872121),
(2.06549358, 16.61084154, 0.0639475 ),
(2.0701642 , 15.69277454, 0.06156768),
(2.07483506, 15.01033761, 0.06108708),
(2.07950568, 14.67329672, 0.06055912),
(2.0841763 , 14.5614213 , 0.05847086),
(2.08884716, 14.3771121 , 0.05891275),
(2.09351778, 14.25279852, 0.05998107),
(2.0981884 , 14.25994032, 0.05832638),
(2.10285926, 14.23462592, 0.05942212),
(2.10752988, 14.20510948, 0.05972571),
(2.1122005 , 14.09332883, 0.0590418 ),
(2.11687136, 14.10938846, 0.05772431),
(2.12154198, 14.21478398, 0.05830313),
(2.1262126 , 14.28427848, 0.0584818 ),
(2.13088346, 14.32668972, 0.05868324),
(2.13555408, 14.35109908, 0.06058188),
(2.14022493, 14.41141221, 0.0654428 ),
(2.14489555, 14.48783474, 0.06164804),
(2.14956617, 14.43695253, 0.05853947),
(2.15423703, 14.53766106, 0.05931266),
(2.15890765, 14.54226979, 0.05933905),
(2.16357827, 14.63735923, 0.06272671),
(2.16824913, 14.59274168, 0.06241335),
(2.17291975, 14.62996471, 0.06227284),
(2.17759037, 14.78774487, 0.06166981),
(2.18226123, 14.87656607, 0.06212647),
(2.18693185, 14.904289 , 0.06553384),
(2.19160271, 14.78106993, 0.06598214),
(2.19627333, 14.35750049, 0.06232894),
(2.20094395, 14.01928977, 0.06014454),
(2.20561481, 13.6717337 , 0.06316704),
(2.21028543, 13.27033976, 0.06272573),
(2.21495605, 13.45858388, 0.06378313),
(2.2196269 , 14.4814341 , 0.07466454),
(2.22429752, 15.33204144, 0.08537209),
(2.22896814, 18.84246093, 0.12252199),
(2.233639 , 20.87433452, 0.17291249),
(2.23830962, 28.3669051 , 0.28658091),
(2.24298024, 31.28393199, 0.3807091 ),
(2.2476511 , 39.62514071, 0.76141747),
(2.25232172, 51.17825988, 1.22953228),
(2.25699258, 63.41328878, 2.06786936)],
dtype=(numpy.record, [('wavelength', '>f8'), ('flux', '>f8'), ('uncertainty', '>f8')]))
# Normalization of observed spectra; Just for visual purpose;
flux_normalize = 500.
wave_200 = fd['wavelength']
flux_200 = fd['flux'] / flux_normalize
flux_err_200 = fd['uncertainty'] / flux_normalize
# Open files for other filters.
filt = 'f150w'
file_1d = f'{DIR_OUT}l3_nis_{filt}_{grism}_s{id}_combine_1d_opt.fits'
fd = fits.open(file_1d)[1].data
wave_150 = fd['wavelength']
flux_150 = fd['flux'] / flux_normalize
flux_err_150 = fd['uncertainty'] / flux_normalize
#
filt = 'f115w'
file_1d = f'{DIR_OUT}l3_nis_{filt}_{grism}_s{id}_combine_1d_opt.fits'
fd = fits.open(file_1d)[1].data
wave_115 = fd['wavelength']
flux_115 = fd['flux'] / flux_normalize
flux_err_115 = fd['uncertainty'] / flux_normalize
# plot all
plt.figure()
plt.errorbar(wave_115, flux_115, ls='-', color='lightblue', label='F115W')
plt.errorbar(wave_150, flux_150, ls='-', color='orange', label='F150W')
plt.errorbar(wave_200, flux_200, ls='-', color='purple', label='F200W')
plt.fill_between(wave_115, flux_115-flux_err_115, flux_115+flux_err_115, ls='-', color='gray', label='', alpha=0.3)
plt.fill_between(wave_150, flux_150-flux_err_150, flux_150+flux_err_150, ls='-', color='gray', label='', alpha=0.3)
plt.fill_between(wave_200, flux_200-flux_err_200, flux_200+flux_err_200, ls='-', color='gray', label='', alpha=0.3)
plt.xlabel('Wavelength / um')
plt.ylim(0., 0.2)
plt.legend(loc=2)
<matplotlib.legend.Legend at 0x7fad2f3c5610>
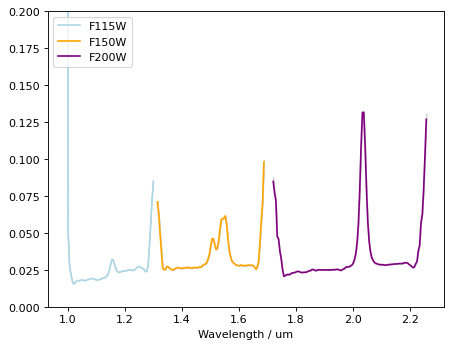
Continuum is visible. We need to subtract it before template correlation.#
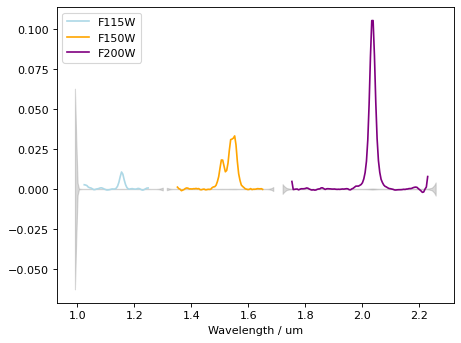
Strong emission lines are seen. (Hb+Oiii doublet at ~1.55um, and blended Ha+Nii at ~2.1um)
Note that flux excess at the edges of each filter are not real, and should be masked out.
# F200W
spec_unit = u.MJy
mask = ((wave_200 > 1.75) & (wave_200 < 1.97)) | ((wave_200 > 2.08) & (wave_200 < 2.23))
obs_200 = Spectrum(spectral_axis=wave_200[mask]*u.um, flux=flux_200[mask]*spec_unit)
continuum_200 = continuum.fit_generic_continuum(obs_200, model=Chebyshev1D(7))
plt.figure()
plt.errorbar(wave_200, flux_200, ls='-', color='purple', label='F200W')
plt.plot(wave_200[mask], continuum_200(obs_200.spectral_axis), color='r', label='Fitted continuum')
plt.legend(loc=0)
flux_200_sub = flux_200 * spec_unit - continuum_200(wave_200*u.um)
mask_200 = (wave_200 > 1.75) & (wave_200 < 2.23)
WARNING: Model is linear in parameters; consider using linear fitting methods. [astropy.modeling.fitting]
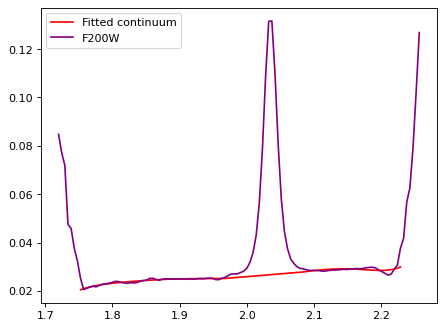
# F150W
spec_unit = u.MJy
mask = ((wave_150 > 1.35) & (wave_150 < 1.48)) | ((wave_150 > 1.6) & (wave_150 < 1.65))
obs_150 = Spectrum(spectral_axis=wave_150[mask]*u.um, flux=flux_150[mask]*spec_unit)
continuum_150 = continuum.fit_generic_continuum(obs_150, model=Chebyshev1D(7))
flux_150_sub = flux_150 * spec_unit - continuum_150(wave_150 * u.um)
plt.figure()
plt.errorbar(wave_150, flux_150, ls='-', color='orange', label='F150W')
plt.plot(wave_150[mask], continuum_150(obs_150.spectral_axis), color='r', label='Fitted continuum')
plt.legend(loc=0)
mask_150 = (wave_150 > 1.35) & (wave_150 < 1.65)
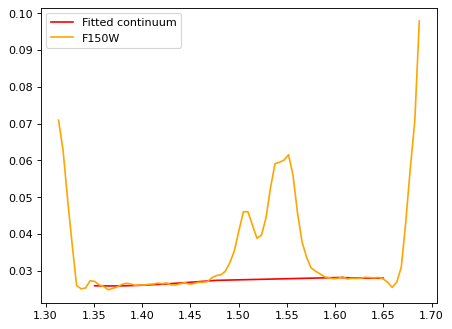
# F150W
spec_unit = u.MJy
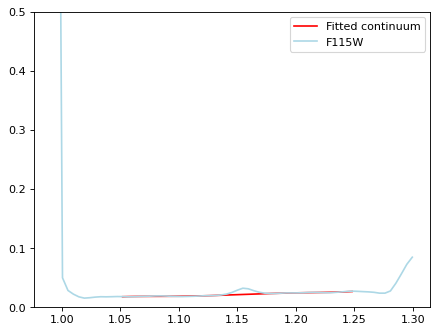
mask = ((wave_115 > 1.05) & (wave_115 < 1.13)) | ((wave_115 > 1.17) & (wave_115 < 1.25))
obs_115 = Spectrum(spectral_axis=wave_115[mask]*u.um, flux=flux_115[mask]*spec_unit)
continuum_115 = continuum.fit_generic_continuum(obs_115, model=Chebyshev1D(7))
plt.figure()
plt.errorbar(wave_115, flux_115, ls='-', color='lightblue', label='F115W')
plt.plot(wave_115[mask], continuum_115(obs_115.spectral_axis), color='r', label='Fitted continuum')
plt.ylim(0, 0.5)
plt.legend(loc=0)
flux_115_sub = flux_115 * spec_unit - continuum_115(wave_115 * u.um)
mask_115 = (wave_115 > 1.02) & (wave_115 < 1.25)
# plot all
plt.figure()
plt.errorbar(wave_115[mask_115], flux_115_sub[mask_115], ls='-', color='lightblue', label='F115W')
plt.errorbar(wave_150[mask_150], flux_150_sub[mask_150], ls='-', color='orange', label='F150W')
plt.errorbar(wave_200[mask_200], flux_200_sub[mask_200], ls='-', color='purple', label='F200W')
plt.fill_between(wave_115, -flux_err_115, flux_err_115, ls='-', color='gray', label='', alpha=0.3)
plt.fill_between(wave_150, -flux_err_150, flux_err_150, ls='-', color='gray', label='', alpha=0.3)
plt.fill_between(wave_200, -flux_err_200, flux_err_200, ls='-', color='gray', label='', alpha=0.3)
plt.xlabel('Wavelength / um')
plt.legend(loc=2)
<matplotlib.legend.Legend at 0x7fad2f189710>
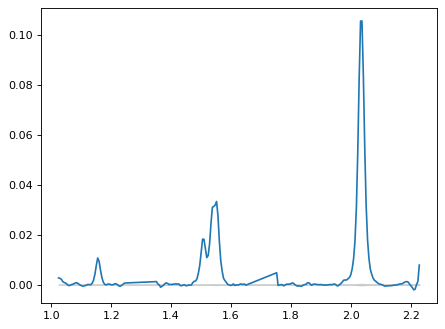
# Concatenate arrays
wave_obs = np.concatenate([wave_115[mask_115], wave_150[mask_150], wave_200[mask_200]])
flux_obs = np.concatenate([flux_115_sub[mask_115], flux_150_sub[mask_150], flux_200_sub[mask_200]])
flux_err_obs = np.concatenate([flux_err_115[mask_115], flux_err_150[mask_150], flux_err_200[mask_200]])
plt.figure()
plt.plot(wave_obs, flux_obs)
plt.fill_between(wave_obs, -flux_err_obs, flux_err_obs, ls='-', color='gray', label='', alpha=0.3)
<matplotlib.collections.FillBetweenPolyCollection at 0x7fad2f2332d0>
wht_obs = 1 / flux_err_obs**2
spec_unit = u.MJy
dataspec = QTable([wave_obs*u.um, flux_obs*spec_unit, wht_obs, flux_err_obs],
names=('wavelength', 'flux', 'weight', 'uncertainty'))
dataspec_sub = dataspec[dataspec['weight'] > 0.]
# Now make it into a Spectrum instance.
p_obs = Spectrum(spectral_axis=dataspec_sub['wavelength'],
flux=dataspec_sub['flux'],
uncertainty=StdDevUncertainty(dataspec_sub['uncertainty']), unit='MJy')
Do cross-correlation with an input template;#
(*Another notebook on Specutils cross correlation by Ivo Busko: https://spacetelescope.github.io/jdat_notebooks/pages/redshift_crosscorr/redshift_crosscorr.html)
Load synthetic template;#
Just to test the functionality, for now we are using the input spectral template used for simulation.
def read_hdf5(filename):
contents = {}
with h5py.File(filename, 'r') as file_obj:
for key in file_obj.keys():
dataset = file_obj[key]
try:
wave_units_string = dataset.attrs['wavelength_units']
except KeyError:
wave_units_string = 'micron'
# Catch common errors
if wave_units_string.lower() in ['microns', 'angstroms', 'nanometers']:
wave_units_string = wave_units_string[0:-1]
# Get the data
waves = dataset[0]
fluxes = dataset[1]
contents[int(key)] = {'wavelengths': waves, 'fluxes': fluxes}
return contents
# Download files, if not exists yet.
if not os.path.exists('./pipeline_products'):
import zipfile
import urllib.request
boxlink = 'https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/NIRISS_lensing_cluster/pipeline_products.zip'
boxfile = './pipeline_products.zip'
urllib.request.urlretrieve(boxlink, boxfile)
zf = zipfile.ZipFile(boxfile, 'r')
list_names = zf.namelist()
for item in list_names:
zf.extract(member=item) # Using extract instead of extractall because it is safer when files have absolute (/) or relative (..) paths
# Template file;
file_temp = './pipeline_products/source_sed_file_from_sources_extend_01_and_sed_file.hdf5'
content = read_hdf5(file_temp)
content
{10177: {'wavelengths': array([3.0999999e-02, 3.1072199e-02, 3.1144397e-02, ..., 3.4099831e+01,
3.4099903e+01, 3.4099976e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.4608030e-25,
8.3700207e-26, 2.1320117e-26], shape=(471877,), dtype=float32)},
10265: {'wavelengths': array([3.0999999e-02, 3.1072199e-02, 3.1144397e-02, ..., 3.4099831e+01,
3.4099903e+01, 3.4099976e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.7282180e-25,
9.9022388e-26, 2.5222983e-26], shape=(471877,), dtype=float32)},
10455: {'wavelengths': array([3.0999999e-02, 3.1072199e-02, 3.1144397e-02, ..., 3.4099831e+01,
3.4099903e+01, 3.4099976e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.4816426e-26,
8.4894260e-27, 2.1624266e-27], shape=(471877,), dtype=float32)},
10709: {'wavelengths': array([3.0999999e-02, 3.1072199e-02, 3.1144397e-02, ..., 3.4099831e+01,
3.4099903e+01, 3.4099976e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 3.6238049e-24,
2.0763458e-24, 5.2888681e-25], shape=(471877,), dtype=float32)},
11151: {'wavelengths': array([3.5000000e-02, 3.5081513e-02, 3.5163030e-02, ..., 3.8499809e+01,
3.8499889e+01, 3.8499973e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.0507161e-24,
6.0203300e-25, 1.5334984e-25], shape=(471877,), dtype=float32)},
11420: {'wavelengths': array([3.5000000e-02, 3.5081513e-02, 3.5163030e-02, ..., 3.8499809e+01,
3.8499889e+01, 3.8499973e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.0168714e-24,
5.8264083e-25, 1.4841028e-25], shape=(471877,), dtype=float32)},
11423: {'wavelengths': array([3.5000000e-02, 3.5081513e-02, 3.5163030e-02, ..., 3.8499809e+01,
3.8499889e+01, 3.8499973e+01], shape=(471877,), dtype=float32),
'fluxes': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 1.0033151e-24,
5.7487341e-25, 1.4643175e-25], shape=(471877,), dtype=float32)}}
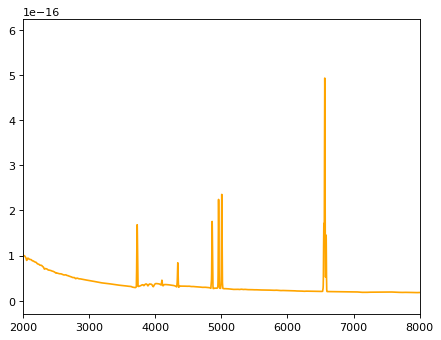
# The template is already redshifted for simulation.
# Now, blueshift it to z=0;
z_input = 2.1
iix = 10709 # Index for the particular template.
flux_all = content[iix]['fluxes']
wave_all = content[iix]['wavelengths'] / (1. + z_input) * 1e4
plt.plot(wave_all, flux_all, color='orange')
plt.xlim(2000, 8000)
(2000.0, 8000.0)
# Cut templates for better fit.
con_tmp = (wave_all > 3600) & (wave_all < 13000)
flux = flux_all[con_tmp]
wave = wave_all[con_tmp]
flux /= flux.max()
wave *= 1.e-4 # force wavelengths in um. (with_spectral_unit doesn't convert)
factor = p_obs.flux.unit # normalize template to a sensible range
template = Spectrum(spectral_axis=wave*u.um, flux=flux*factor)
plt.figure()
plt.plot(template.spectral_axis, template.flux, color='orange')
plt.xlabel(template.spectral_axis.unit)
Text(0.5, 0, 'um')
Binning the template into a course array, if you have too many elements and cause an issue below;#
-> In this example this is not necessary.#
# Make this true, if you want to bin;
if False:
wave_bin = np.arange(wave.min(), wave.max(), 20.0)
flux_bin = np.interp(wave_bin, wave, flux)
else:
wave_bin = wave
flux_bin = flux
factor = p_obs.flux.unit # normalize template to a sensible range
template = Spectrum(spectral_axis=wave_bin*u.um, flux=flux_bin*factor)
plt.figure()
plt.plot(template.spectral_axis, template.flux, color='orange')
plt.xlabel(template.spectral_axis.unit)
Text(0.5, 0, 'um')

# Continuum fit to the fitting temp;
regions = [SpectralRegion(0.37*u.um, 0.452*u.um),
SpectralRegion(0.476*u.um, 0.53*u.um),
SpectralRegion(0.645*u.um, 0.67*u.um)]
continuum_model = continuum.fit_generic_continuum(template, model=Chebyshev1D(3), exclude_regions=regions)
p_template = template - continuum_model(template.spectral_axis)
plt.figure()
plt.plot(p_template.spectral_axis, p_template.flux, color='orange')
plt.xlabel(p_template.spectral_axis.unit)
plt.title('Continuum subtracted template')
Text(0.5, 1.0, 'Continuum subtracted template')
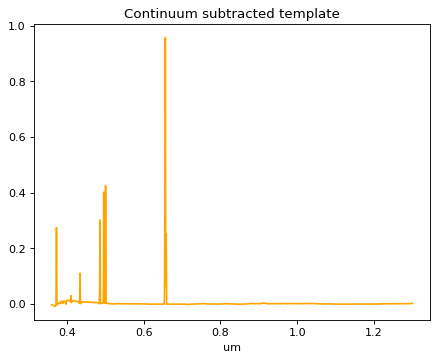
# Put data array into Specutils format;
sflux = p_template.flux
p_smooth_template = Spectrum(spectral_axis=p_template.spectral_axis,
flux=sflux/sflux.max())
plt.figure()
plt.plot(p_smooth_template.spectral_axis, p_smooth_template.flux, color='b', label='Smoothed template')
plt.plot(p_obs.spectral_axis, p_obs.flux, color='red', label='Spectrum')
plt.xlabel(p_smooth_template.spectral_axis.unit)
plt.legend(loc=0)
<matplotlib.legend.Legend at 0x7fad2cf67f10>
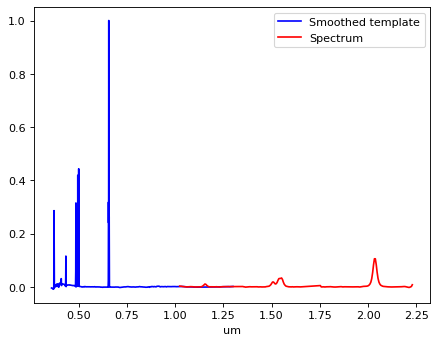
CAUTION: Two inputs spectra above have to overlap, if only a small part, to make Specutils works properly.#
Cross Correlation using specutils functionarity;#
# With no additional specifications, both the entire template and entire spectrum
# will be included in the correlation computation. This in general will incur in
# a significant increase in execution time. It is advised that the template is cut
# to work only on the useful region.
corr, lag = correlation.template_correlate(p_obs, p_smooth_template)
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/units/quantity.py:648: RuntimeWarning: invalid value encountered in divide
result = super().__array_ufunc__(function, method, *arrays, **kwargs)
plt.figure()
plt.gcf().set_size_inches((8., 4.))
plt.plot(lag, corr, linewidth=0.5)
plt.xlabel(lag.unit)
Text(0.5, 0, 'km / s')
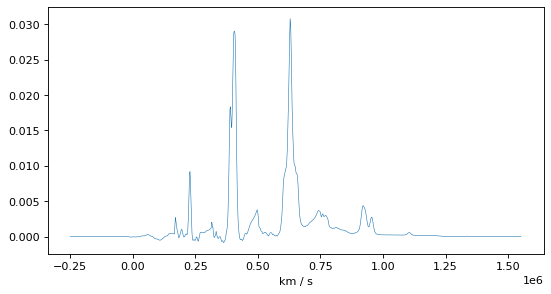
# Redshift based on maximum
index_peak = np.argmax(corr)
v = lag[index_peak]
z = v / const.c.to('km/s')
print("Peak maximum at: ", v)
print("Redshift from peak maximum: ", z)
Peak maximum at: 628833.1938062641 km / s
Redshift from peak maximum: 2.0975617532255066
# Redshift based on parabolic fit to maximum
n = 8 # points to the left or right of correlation maximum
peak_lags = lag[index_peak-n:index_peak+n+1].value
peak_vals = corr[index_peak-n:index_peak+n+1].value
p = np.polyfit(peak_lags, peak_vals, deg=2)
roots = np.roots(p)
v_fit = np.mean(roots) * u.km/u.s # maximum lies at mid point between roots
z = v_fit / const.c.to('km/s')
print("")
print("Parabolic fit with maximum at: ", v_fit)
print("Redshift from parabolic fit: ", z)
Parabolic fit with maximum at: 629197.8938783638 km / s
Redshift from parabolic fit: 2.098778261721193
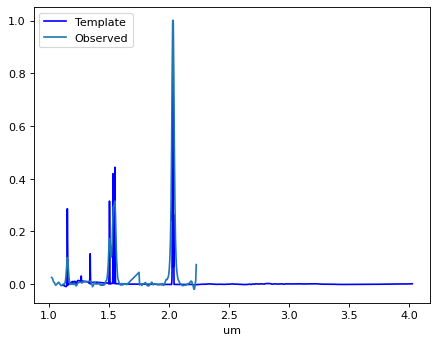
How does it look?#
print('z =', z)
plt.figure()
plt.plot(
template.spectral_axis * (1. + z),
p_smooth_template.flux / np.max(p_smooth_template.flux),
color='b', label='Template'
)
plt.plot(p_obs.spectral_axis, p_obs.flux / np.max(p_obs.flux), label='Observed')
plt.legend(loc=2)
plt.xlabel(p_obs.spectral_axis.unit)
z = 2.098778261721193
Text(0.5, 0, 'um')