MAST Table Access Protocol PanSTARRS 1 DR2 Demo#
This tutorial demonstrates how to use astroquery to access PanSTARRS 1 Data Release 2 via a Virtual Observatory standard Table Access Protocol (TAP) service at MAST, and work with the resultant data. It relies on Python 3 and astroquery, as well as some other common scientific packages.
Table of Contents#
TAP Service Introduction#
Table Access Protocol (TAP) services allow more direct and flexible access to astronomical data than the simpler types of IVOA standard data services. Queries are built with the SQL-like Astronomical Data Query Language (ADQL), and can include geographic / spatial queries as well as filtering on other characteristics of the data. This also allows the user fine-grained control over the returned columns, unlike the fixed set of coumns returned from cone, image, and spectral services.
For this example, we’ll be using the astropy affiliated PyVO client, which is interoperable with other valid TAP services, including those at MAST. PyVO documentation is available at ReadTheDocs: https://pyvo.readthedocs.io
We’ll be using PyVO to call the TAP service at MAST serving PanSTARRS 1 Data Release 2, now with individual detection information. The schema is described within the service, and we’ll show how to inspect it. The schema is also the same as the one available via the CasJobs interface, with an additional view added for the most common positional queries. CasJobs has its own copy of the schema documentation, which can be accessed through its own site: http://mastweb.stsci.edu/ps1casjobs/
Imports#
# Use the pyvo library as our client to the data service.
import pyvo as vo
# For resolving objects with tools from MAST
from astroquery.mast import Mast
# For displaying and manipulating some types of results
%matplotlib inline
import numpy as np
import pylab
# suppress unimportant unit warnings from many TAP services
import warnings
warnings.filterwarnings("ignore", module="astropy.io.votable.*")
warnings.filterwarnings("ignore", module="pyvo.utils.xml.elements")
Connecting to a TAP Service#
The PyVO library is able to connect to any TAP service, given the “base” URL as noted in metadata registry resources describing the service. This is the URL for the PanSTARRS 1 DR2 TAP service.
TAP_service = vo.dal.TAPService("https://mast.stsci.edu/vo-tap/api/v0.1/ps1dr2/")
TAP_service.describe()
Capability ivo://ivoa.net/std/TAP
Interface vs:ParamHTTP
https://mast.stsci.edu/vo-tap/api/v0.1/ps1dr2
Language ADQL
ivo://ivoa.net/std/TAPRegExt#features-adqlgeo
CONTAINS
POINT
CIRCLE
Output format application/json
Also available as json
Output format text/csv;header=present
Also available as csv
Output format application/xml
Also available as xml
Output format application/x-votable+xml
Also available as votable
Maximum size of resultsets
Default 100000 row
Maximum 100000 row
Capability ivo://ivoa.net/std/DALI#examples
Interface vr:WebBrowser
https://mast.stsci.edu/vo-tap/api/v0.1/ps1dr2/examples
List available tables#
There are MANY tables available, so we’ll only print out the first few. Use the commented-out for loop to see them all.
TAP_tables = TAP_service.tables
# only show the first three tables
for i, tablename in enumerate(TAP_tables.keys()):
if i < 10 and "tap_schema" not in tablename:
TAP_tables[tablename].describe()
print("Columns={}".format(sorted([k.name for k in TAP_tables[tablename].columns])))
print("----")
# PRINT ALL TABLES
# for tablename in TAP_tables.keys():
# if not "tap_schema" in tablename:
# TAP_tables[tablename].describe()
# print("Columns={}".format(sorted([k.name for k in TAP_tables[tablename].columns ])))
# print("----")
WARNING: W02: None:1:1760: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:1974: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:13049: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
dbo.detection
Contains single epoch photometry of individual detections from a single exposure. The identifiers connecting the detection back to the original image and to the object association are provided. PSF; aperture; and Kron (1980) photometry are included; along with sky and detector coordinate positions. References: Kron; R. G. 1980; ApJS; 43; 305.
Columns=['airmass', 'apfillf', 'apflux', 'apfluxerr', 'apradius', 'dec', 'decerr', 'detectid', 'dvoregionid', 'exptime', 'extnsigma', 'filterid', 'imageid', 'infoflag', 'infoflag2', 'infoflag3', 'ippdetectid', 'ippobjid', 'kronflux', 'kronfluxerr', 'kronrad', 'momentm3c', 'momentm3s', 'momentm4c', 'momentm4s', 'momentr1', 'momentrh', 'momentxx', 'momentxy', 'momentyy', 'objid', 'obstime', 'pltscale', 'posangle', 'processingversion', 'psfchisq', 'psfcore', 'psfflux', 'psffluxerr', 'psflikelihood', 'psfmajorfwhm', 'psfminorfwhm', 'psfqf', 'psfqfperfect', 'psftheta', 'ra', 'raerr', 'randomdetid', 'sky', 'skyerr', 'surveyid', 'telluricext', 'uniquepspsp2id', 'xpos', 'xposerr', 'ypos', 'yposerr', 'zp']
----
dbo.detectionflags
Contains information flag values for detections.
Columns=['"value"', 'description', 'hexadecimal', 'name']
----
dbo.detectionflags2
Contains information flag values for detections. References: Petrosian; V. 1976; ApJL; 209; L1.
Columns=['"value"', 'description', 'hexadecimal', 'name']
----
dbo.detectionflags3
Contains information flag values for detections. References: Kron; R. G. 1980; ApJS; 43; 305.
Columns=['"value"', 'description', 'hexadecimal', 'name']
----
dbo.detectionobjectview
-- ObjectThin join Detection joined by objID column.
Columns=['airmass', 'apfillf', 'apflux', 'apfluxerr', 'apradius', 'b', 'batchid', 'beta', 'cx', 'cy', 'cz', 'dec', 'decerr', 'decmean', 'decmeanerr', 'decstack', 'decstackerr', 'detectid', 'dvoregionid', 'epochmean', 'exptime', 'extnsigma', 'filterid', 'gflags', 'gmeanapmag', 'gmeanapmagerr', 'gmeanapmagnpt', 'gmeanapmagstd', 'gmeankronmag', 'gmeankronmagerr', 'gmeankronmagnpt', 'gmeankronmagstd', 'gmeanpsfmag', 'gmeanpsfmagerr', 'gmeanpsfmagmax', 'gmeanpsfmagmin', 'gmeanpsfmagnpt', 'gmeanpsfmagstd', 'gqfperfect', 'htmid', 'iflags', 'imageid', 'imeanapmag', 'imeanapmagerr', 'imeanapmagnpt', 'imeanapmagstd', 'imeankronmag', 'imeankronmagerr', 'imeankronmagnpt', 'imeankronmagstd', 'imeanpsfmag', 'imeanpsfmagerr', 'imeanpsfmagmax', 'imeanpsfmagmin', 'imeanpsfmagnpt', 'imeanpsfmagstd', 'infoflag', 'infoflag2', 'infoflag3', 'ippobjid', 'iqfperfect', 'kronflux', 'kronfluxerr', 'kronrad', 'l', 'lambda', 'momentm3c', 'momentm3s', 'momentm4c', 'momentm4s', 'momentr1', 'momentrh', 'momentxx', 'momentxy', 'momentyy', 'ndetections', 'ng', 'ni', 'nr', 'nstackdetections', 'nstackobjectrows', 'ny', 'nz', 'objaltname1', 'objaltname2', 'objaltname3', 'objid', 'objinfoflag', 'objname', 'obstime', 'pltscale', 'posangle', 'posmeanchisq', 'processingversion', 'projectionid', 'psfchisq', 'psfcore', 'psfflux', 'psffluxerr', 'psflikelihood', 'psfmajorfwhm', 'psfminorfwhm', 'psfqf', 'psfqfperfect', 'psftheta', 'qualityflag', 'ra', 'raerr', 'ramean', 'rameanerr', 'randomdetid', 'randomid', 'rastack', 'rastackerr', 'rflags', 'rmeanapmag', 'rmeanapmagerr', 'rmeanapmagnpt', 'rmeanapmagstd', 'rmeankronmag', 'rmeankronmagerr', 'rmeankronmagnpt', 'rmeankronmagstd', 'rmeanpsfmag', 'rmeanpsfmagerr', 'rmeanpsfmagmax', 'rmeanpsfmagmin', 'rmeanpsfmagnpt', 'rmeanpsfmagstd', 'rqfperfect', 'sky', 'skycellid', 'skyerr', 'surveyid', 'telluricext', 'tessid', 'uniquepspsobid', 'uniquepspsp2id', 'xpos', 'xposerr', 'yflags', 'ymeanapmag', 'ymeanapmagerr', 'ymeanapmagnpt', 'ymeanapmagstd', 'ymeankronmag', 'ymeankronmagerr', 'ymeankronmagnpt', 'ymeankronmagstd', 'ymeanpsfmag', 'ymeanpsfmagerr', 'ymeanpsfmagmax', 'ymeanpsfmagmin', 'ymeanpsfmagnpt', 'ymeanpsfmagstd', 'ypos', 'yposerr', 'yqfperfect', 'zflags', 'zmeanapmag', 'zmeanapmagerr', 'zmeanapmagnpt', 'zmeanapmagstd', 'zmeankronmag', 'zmeankronmagerr', 'zmeankronmagnpt', 'zmeankronmagstd', 'zmeanpsfmag', 'zmeanpsfmagerr', 'zmeanpsfmagmax', 'zmeanpsfmagmin', 'zmeanpsfmagnpt', 'zmeanpsfmagstd', 'zoneid', 'zp', 'zqfperfect']
----
WARNING: W02: None:1:5423: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:21625: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:24170: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:30420: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:30820: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
WARNING: W02: None:1:31393: W02: 'unsignedbyte' is not a valid datatype according to the VOSI spec [pyvo.io.vosi.vodataservice]
Use Cases#
Simple Positional Query#
This searches the mean object catalog for objects within .2 degrees of M87 (RA=187.706, Dec=12.391 in degrees). The view used contains information from the ObjectThin table (which has information on object positions and the number of available measurements) and the MeanObject table (which has information on photometry averaged over the multiple epochs of observation).
Note that the results are restricted to objects with nDetections>1, where nDetections is the total number of times the object was detected on the single-epoch images in any filter at any time. Objects with nDetections=1 tend to be artifacts, so this is a quick way to eliminate most spurious objects from the catalog.
This query runs in TAP’s asynchronous mode, which is a queued batch mode with some overhead and longer timeouts, useful for big catalogs like PanSTARRS. It may not be necessary for all queries to PS1 DR2, but the PyVO client can automatically handle the additional processing required over synchronous mode.
job = TAP_service.run_async("""
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag, rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',187.706,12.391,.2))=1
AND nDetections > 1
""")
TAP_results = job.to_table()
TAP_results
| objid | ramean | decmean | ndetections | ng | nr | ni | nz | ny | gmeanpsfmag | rmeanpsfmag | imeanpsfmag | zmeanpsfmag | ymeanpsfmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg | deg | mag | mag | mag | mag | mag | |||||||
| int64 | float64 | float64 | int16 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 |
| 122861877059169881 | 187.70591003309602 | 12.39113345997396 | 54 | 10 | 10 | 14 | 8 | 12 | 10.295 | 10.732 | 11.076 | 11.342 | 11.557 |
| 122861877056308967 | 187.70559948 | 12.39030211 | 2 | 0 | 0 | 0 | 0 | 2 | -999.0 | -999.0 | -999.0 | -999.0 | 14.9985 |
| 122871877063310741 | 187.70631118732143 | 12.39181079630862 | 3 | 0 | 2 | 0 | 1 | 0 | -999.0 | 14.4229 | -999.0 | 14.4249 | -999.0 |
| 122861877058698594 | 187.70580919 | 12.39013311 | 2 | 0 | 0 | 0 | 0 | 2 | -999.0 | -999.0 | -999.0 | -999.0 | 14.7305 |
| 122861877050678994 | 187.70512542608552 | 12.390430557059906 | 5 | 2 | 2 | 1 | 0 | 0 | 17.0287 | 15.0843 | 16.169 | -999.0 | -999.0 |
| 122871877062551436 | 187.70622365 | 12.39236505 | 2 | 0 | 0 | 0 | 0 | 2 | -999.0 | -999.0 | -999.0 | -999.0 | 15.1102 |
| 122861877044629638 | 187.70451598 | 12.39086724 | 2 | 0 | 0 | 0 | 2 | 0 | -999.0 | -999.0 | -999.0 | 15.5294 | -999.0 |
| 122861877056688054 | 187.70558996 | 12.38960002 | 2 | 0 | 0 | 0 | 2 | 0 | -999.0 | -999.0 | -999.0 | 14.4064 | -999.0 |
| 122871877075050430 | 187.70747912 | 12.3915839 | 2 | 0 | 0 | 0 | 2 | 0 | -999.0 | -999.0 | -999.0 | 15.5752 | -999.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 122901875031002013 | 187.50314518107095 | 12.417915440212205 | 37 | 7 | 10 | 11 | 7 | 2 | 21.2326 | 20.7473 | 20.5742 | 20.6803 | 20.2113 |
| 122661878055210091 | 187.80550736264715 | 12.216281397817458 | 57 | 13 | 14 | 12 | 6 | 12 | 20.9059 | 20.3516 | 20.0699 | 19.9956 | 19.8391 |
| 122631877536376320 | 187.75369336 | 12.19655761 | 2 | 0 | 0 | 0 | 2 | 0 | -999.0 | -999.0 | -999.0 | 21.0599 | -999.0 |
| 123031875562633406 | 187.55622393040358 | 12.527354381090412 | 3 | 0 | 0 | 3 | 0 | 0 | -999.0 | -999.0 | 20.5518 | -999.0 | -999.0 |
| 122681878343382931 | 187.83438155722584 | 12.235251674424532 | 24 | 5 | 9 | 6 | 4 | 0 | 22.0912 | 21.7042 | 21.3068 | 21.0164 | -999.0 |
| 123061878259604285 | 187.82588872 | 12.55313763 | 2 | 1 | 0 | 1 | 0 | 0 | 21.9049 | -999.0 | 21.4564 | -999.0 | -999.0 |
| 122751878844752179 | 187.88446793636015 | 12.293022278414929 | 77 | 16 | 24 | 14 | 11 | 12 | 21.4623 | 20.3555 | 20.1394 | 20.0368 | 19.8008 |
| 123001875376856622 | 187.53785086789503 | 12.505173082359276 | 4 | 0 | 3 | 1 | 0 | 0 | -999.0 | 21.1887 | 21.1419 | -999.0 | -999.0 |
| 123101876640214622 | 187.66398717765725 | 12.586746063373354 | 3 | 0 | 0 | 2 | 1 | 0 | -999.0 | -999.0 | 21.8531 | 21.6726 | -999.0 |
Get DR2 light curve for RR Lyrae star KQ UMa#
This time we start with the object name, use the MAST name resolver (which relies on Simbad and NED) to convert the name to RA and Dec, and then query the PS1 DR2 mean object catalog at that position. Then we run a spatial query to TAP using those coordinates.
objname = 'KQ UMa'
coords = Mast.resolve_object(objname)
ra, dec = coords.ra.value, coords.dec.value
radius = 1.0/3600.0 # radius = 1 arcsec
query = """
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag,
rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',{},{},{}))=1
AND nDetections > 1
""".format(ra, dec, radius)
print(query)
job = TAP_service.run_async(query)
TAP_results = job.to_table()
TAP_results
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag,
rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',139.33446271609,68.63508880829,0.0002777777777777778))=1
AND nDetections > 1
| objid | ramean | decmean | ndetections | ng | nr | ni | nz | ny | gmeanpsfmag | rmeanpsfmag | imeanpsfmag | zmeanpsfmag | ymeanpsfmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg | deg | mag | mag | mag | mag | mag | |||||||
| int64 | float64 | float64 | int16 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 |
| 190361393344112894 | 139.33445305334158 | 68.63505916169231 | 66 | 8 | 10 | 21 | 13 | 14 | 15.0402 | 14.553 | 14.2109 | 14.2814 | 14.3041 |
Get Repeated Detection Information#
Extract all the objects with the same object ID from the Detection table, which contains all the individual measurements for this source. The results are joined to the Filter table to convert the filter numbers to names.
objid = TAP_results['objid'][0]
query = """
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID={}
ORDER BY filterID, obsTime
""".format(objid)
print(query)
job = TAP_service.run_async(query)
detection_TAP_results = job.to_table()
detection_TAP_results
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID=190361393344112894
ORDER BY filterID, obsTime
| objID | detectID | filterid | filterType | obsTime | ra | dec | psfFlux | psfFluxErr | psfMajorFWHM | psfMinorFWHM | psfQfPerfect | apFlux | apFluxErr | infoFlag | infoFlag2 | infoFlag3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d | deg | deg | arcsec | arcsec | ||||||||||||
| int64 | int64 | uint8 | object | float64 | float64 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int64 | int32 | int32 |
| 190361393344112894 | 153347716310000010 | 1 | g | 55634.477414 | 139.3345207 | 68.63503577 | 0.00826192 | 1.14074e-05 | 1.88185 | 1.76734 | 0.992916 | 0.00861705 | 1.14233e-05 | 102760517 | 128 | 124782656 |
| 190361393344112894 | 153348968310000008 | 1 | g | 55634.4899457 | 139.33448821 | 68.63506146 | 0.0077373 | 1.10268e-05 | 1.81031 | 1.60518 | 0.998461 | 0.00792172 | 1.09406e-05 | 102760517 | 128 | 124782656 |
| 190361393344112894 | 232228791560000017 | 1 | g | 56423.2881719 | 139.33449441 | 68.63503952 | 0.00335198 | 7.26745e-06 | 1.60204 | 1.45048 | 0.998589 | 0.00338658 | 7.2363e-06 | 102760517 | 128 | 108038208 |
| 190361393344112894 | 255559866370000015 | 1 | g | 56656.5989211 | 139.33444592 | 68.63504589 | 0.00372909 | 7.37688e-06 | 1.82831 | 1.68692 | 0.999191 | 0.00372163 | 7.52088e-06 | 102760517 | 128 | 124815424 |
| 190361393344112894 | 262040070370000016 | 1 | g | 56721.4009635 | 139.33445148 | 68.63504807 | 0.003576 | 7.22894e-06 | 1.60585 | 1.50396 | 0.998605 | 0.00359603 | 7.43463e-06 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 262040708370000014 | 1 | g | 56721.4073411 | 139.33445304 | 68.63504406 | 0.00350554 | 7.14809e-06 | 1.62415 | 1.48827 | 0.999487 | 0.00359624 | 7.43434e-06 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 264231864260000022 | 1 | g | 56743.3189045 | 139.3344431 | 68.63504497 | 0.00343867 | 7.81574e-06 | 1.53957 | 1.4761 | 0.999049 | 0.00361239 | 7.85237e-06 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 264232516260000024 | 1 | g | 56743.3254198 | 139.33444512 | 68.63505285 | 0.00347403 | 7.87789e-06 | 1.52751 | 1.43325 | 0.999367 | 0.00360349 | 7.8686e-06 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 153441340410000012 | 2 | r | 55635.4136426 | 139.33447173 | 68.6350534 | 0.00978758 | 1.10843e-05 | 1.67879 | 1.5653 | 0.998509 | 0.00990763 | 1.11231e-05 | 102760517 | 128 | 124815424 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 190361393344112894 | 183255163520000015 | 5 | y | 55933.5518128 | 139.33444232 | 68.63505669 | 0.00915573 | 2.64422e-05 | 1.11331 | 0.95368 | 0.997115 | 0.00914515 | 2.48501e-05 | 102760517 | 128 | 74483776 |
| 190361393344112894 | 191726725200000011 | 5 | y | 56018.2674345 | 139.33446079 | 68.63506403 | 0.00829646 | 2.59461e-05 | 1.07332 | 1.01563 | 0.997924 | 0.00833759 | 2.33303e-05 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 191727369200000012 | 5 | y | 56018.2738664 | 139.33445189 | 68.63506152 | 0.00819996 | 2.55524e-05 | 1.09504 | 1.05914 | 0.997819 | 0.00820753 | 2.3153e-05 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 229122569150000014 | 5 | y | 56392.2258719 | 139.33442742 | 68.63506234 | 0.00780422 | 2.57689e-05 | 1.02878 | 0.893195 | 0.998618 | 0.00777279 | 2.16455e-05 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 229123273150000011 | 5 | y | 56392.2329084 | 139.33444595 | 68.63505744 | 0.00803503 | 2.55974e-05 | 0.941801 | 0.888788 | 0.99805 | 0.00807037 | 2.2092e-05 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 264322648150000024 | 5 | y | 56744.2269452 | 139.334454 | 68.63505864 | 0.00540446 | 1.32146e-05 | 1.38829 | 1.3477 | 0.998922 | 0.00549867 | 1.13527e-05 | 102760517 | 128 | 124782656 |
| 190361393344112894 | 265623008150000027 | 5 | y | 56757.2305455 | 139.33444319 | 68.63506375 | 0.00636614 | 1.60784e-05 | 0.895041 | 0.803365 | 0.852516 | 0.00412859 | 1.01303e-05 | 102760581 | 128 | 34880 |
| 190361393344112894 | 287165374630000023 | 5 | y | 56972.6539243 | 139.33444226 | 68.63506087 | 0.00687817 | 2.79326e-05 | 1.53686 | 1.28813 | 0.991647 | 0.00699914 | 2.16585e-05 | 102760517 | 128 | 7374912 |
| 190361393344112894 | 287265116630000028 | 5 | y | 56973.6513438 | 139.33444991 | 68.63505759 | 0.00665377 | 2.7704e-05 | 1.85619 | 1.70397 | 0.99864 | 0.00663106 | 2.13156e-05 | 102760517 | 128 | 7342144 |
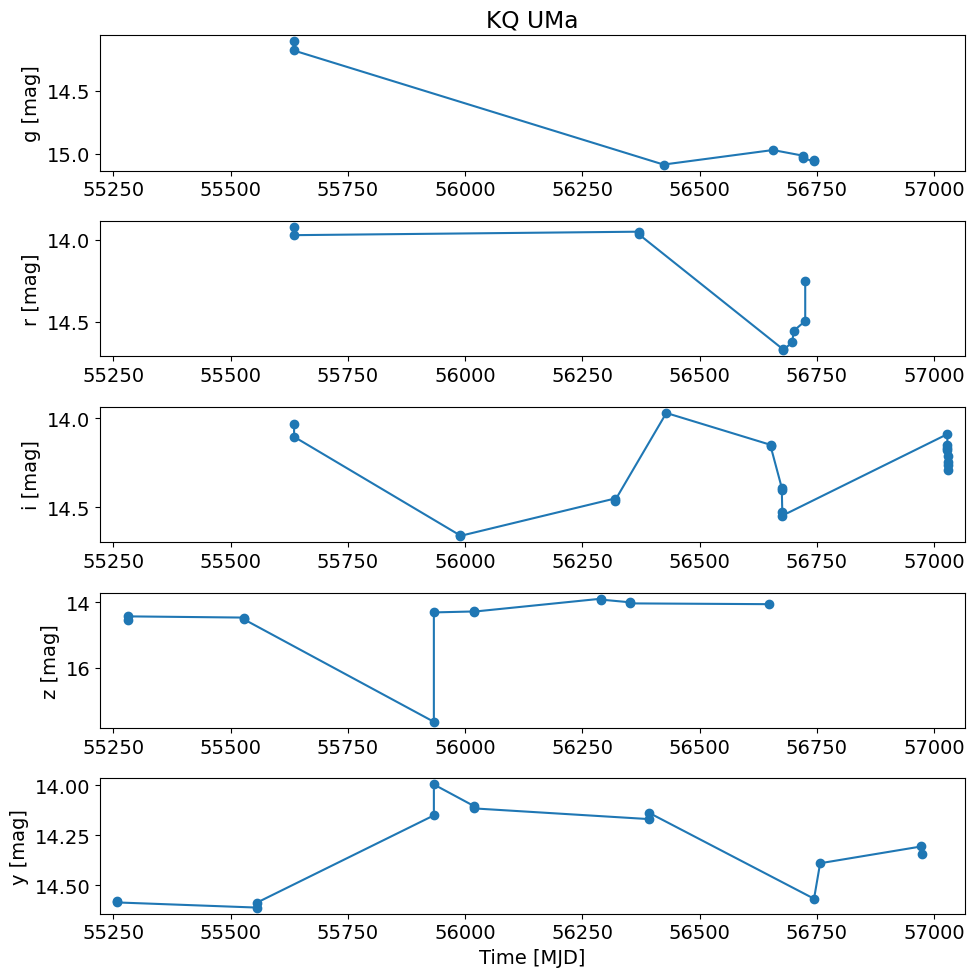
Plot the light curves#
The psfFlux values from the Detection table are converted from Janskys to AB magnitudes. Measurements in the 5 different filters are plotted separately.
# convert flux in Jy to magnitudes
t = detection_TAP_results['obsTime']
mag = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
# detection_TAP_results['filterType'] is a byte string, compare accordingly:
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where(detection_TAP_results['filterType'] == filter)
pylab.plot(t[w], mag[w], '-o')
pylab.ylabel(f'{filter} [mag]')
pylab.xlim(xlim)
pylab.gca().invert_yaxis()
if i == 0:
pylab.title(objname)
pylab.xlabel('Time [MJD]')
pylab.tight_layout()
Plot differences from the mean magnitudes in the initial search.
# convert flux in Jy to magnitudes
t = detection_TAP_results['obsTime']
mag = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
# detection_TAP_results['filterType'] is a byte string, compare accordingly:
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where(detection_TAP_results['filterType'] == filter)
magmean = TAP_results[f'{filter}meanpsfmag'][0]
pylab.plot(t[w], mag[w] - magmean, '-o')
pylab.ylabel(f'{filter} [mag - {np.round(float(magmean), 4)}]')
pylab.xlim(xlim)
pylab.gca().invert_yaxis()
if i == 0:
pylab.title(objname)
pylab.xlabel('Time [MJD]')
pylab.tight_layout()
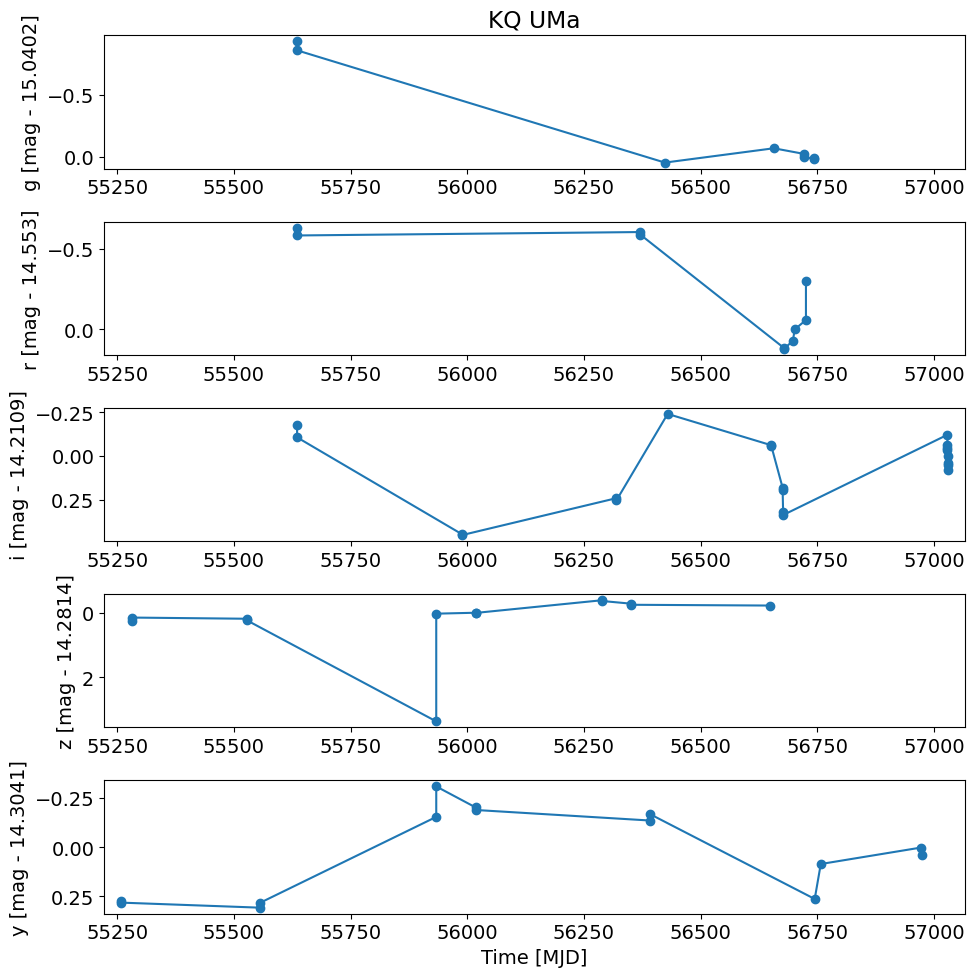
Identify bad data#
There is one clearly bad \(z\) magnitude with a very large difference. Select the bad point and look at it in more detail.
Note that indexing a table (or numpy array) with a logical expression selects just the rows where that expression is true.
detection_TAP_results[(detection_TAP_results['filterType'] == 'z') & (np.abs(mag-TAP_results['zmeanpsfmag'][0]) > 2)]
| objID | detectID | filterid | filterType | obsTime | ra | dec | psfFlux | psfFluxErr | psfMajorFWHM | psfMinorFWHM | psfQfPerfect | apFlux | apFluxErr | infoFlag | infoFlag2 | infoFlag3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d | deg | deg | arcsec | arcsec | ||||||||||||
| int64 | int64 | uint8 | object | float64 | float64 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int64 | int32 | int32 |
| 190361393344112894 | 183252627520000234 | 4 | z | 55933.5264577 | 139.33488168 | 68.63532273 | 0.000317945 | 6.73008e-06 | 1.07537 | 1.0153 | 0.322986 | 0.000213217 | 2.36939e-06 | 102760453 | 128 | 32768 |
From examining this table, it looks like psfQfPerfect is bad. This flag is the PSF-weighted fraction of unmasked pixels in the image (see the documentation for more details). Values near unity indicate good data that is not significantly affected by bad pixels.
Check all the psfQfPerfect values for the \(z\) filter to see if this value really is unusual. The list of values below are sorted by magnitude. The bad point is the only value with psfQfPerfect < 0.95.
w = np.where(detection_TAP_results['filterType'] == 'z')
zdtab = detection_TAP_results[w]
zdtab['mag'] = mag[w]
zdtab['dmag'] = zdtab['mag'] - TAP_results['zmeanpsfmag'][0]
ii = np.argsort(-np.abs(zdtab['dmag']))
zdtab = zdtab[ii]
zdtab['objID', 'obsTime', 'mag', 'dmag', 'psfQfPerfect']
| objID | obsTime | mag | dmag | psfQfPerfect |
|---|---|---|---|---|
| d | ||||
| int64 | float64 | float64 | float64 | float32 |
| 190361393344112894 | 55933.5264577 | 17.644120001792906 | 3.3627202749252305 | 0.322986 |
| 190361393344112894 | 56289.6159346 | 13.890659356117249 | -0.3907403707504269 | 0.997811 |
| 190361393344112894 | 56289.6241112 | 13.91680612564087 | -0.3645936012268063 | 0.988369 |
| 190361393344112894 | 56351.4168483 | 13.998972916603089 | -0.28242681026458705 | 0.999257 |
| 190361393344112894 | 55281.2528285 | 14.537914538383484 | 0.25651481151580846 | 0.99754 |
| 190361393344112894 | 56351.424076 | 14.032501721382141 | -0.2488980054855343 | 0.999187 |
| 190361393344112894 | 55527.6508919 | 14.512118244171143 | 0.23071851730346715 | 0.997265 |
| 190361393344112894 | 56648.5676019 | 14.056430006027222 | -0.22496972084045375 | 0.997982 |
| 190361393344112894 | 55527.6381469 | 14.465140843391419 | 0.18374111652374303 | 0.99738 |
| 190361393344112894 | 55281.2625151 | 14.42855372428894 | 0.147153997421265 | 0.955584 |
| 190361393344112894 | 55933.534515 | 14.307639741897583 | 0.026240015029907582 | 0.997489 |
| 190361393344112894 | 56019.2968782 | 14.278636121749878 | -0.0027636051177974963 | 0.997654 |
| 190361393344112894 | 56019.3038014 | 14.282891297340393 | 0.0014915704727176404 | 0.997338 |
Repeat the plot with bad psfQfPerfect values excluded#
Do the plot again but exclude low psfQfPerfect values.
# convert flux in Jy to magnitudes
t = detection_TAP_results['obsTime']
mag = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
magmean = 0.0*mag
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
magmean[detection_TAP_results['filterType'] == filter] = TAP_results[f'{filter}meanpsfmag'][0]
dmag = mag - magmean
dmag1 = dmag[detection_TAP_results['psfQfPerfect'] > 0.9]
# fix the x and y axis ranges
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
# flip axis direction for magnitude
ylim = np.array([dmag1.max(), dmag1.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where((detection_TAP_results['filterType'] == filter) & (detection_TAP_results['psfQfPerfect'] > 0.9))[0]
pylab.plot(t[w], dmag[w], '-o')
pylab.ylabel('{} [mag - {:.2f}]'.format(filter, magmean[w[0]]))
pylab.xlim(xlim)
pylab.ylim(ylim)
if i == 0:
pylab.title(objname)
pylab.xlabel('Time [MJD]')
pylab.tight_layout()
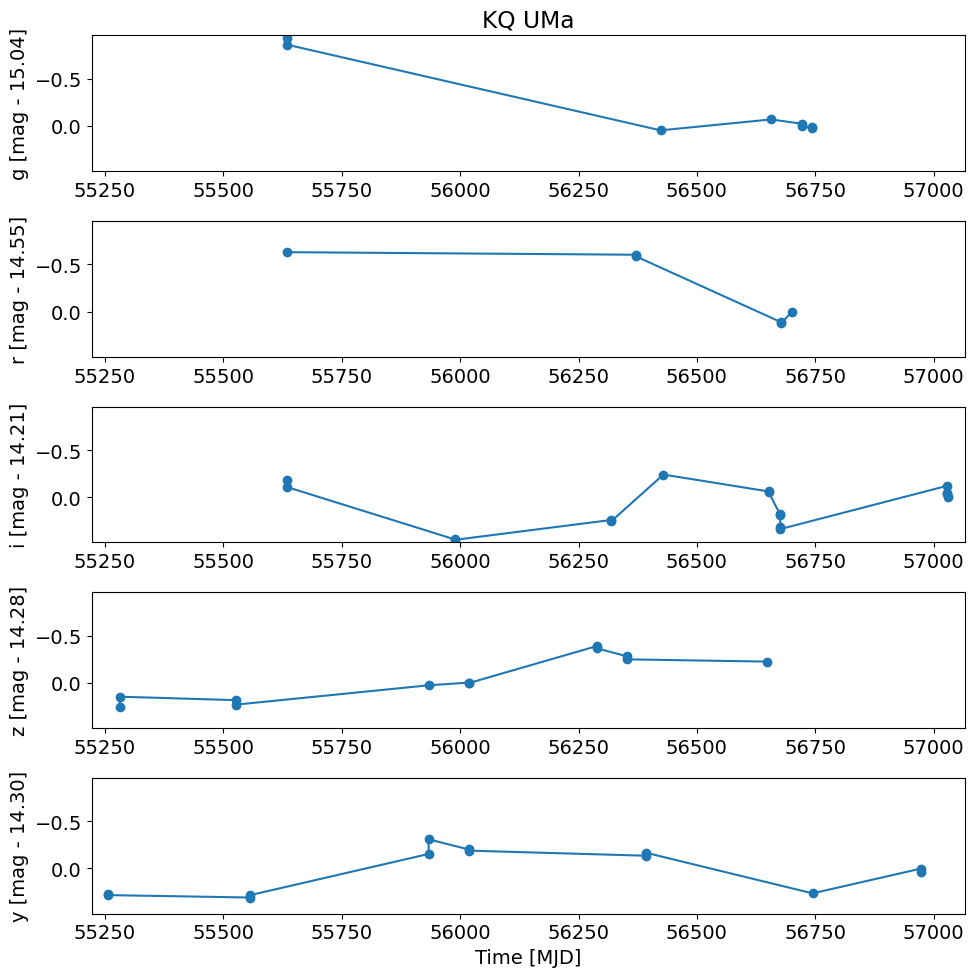
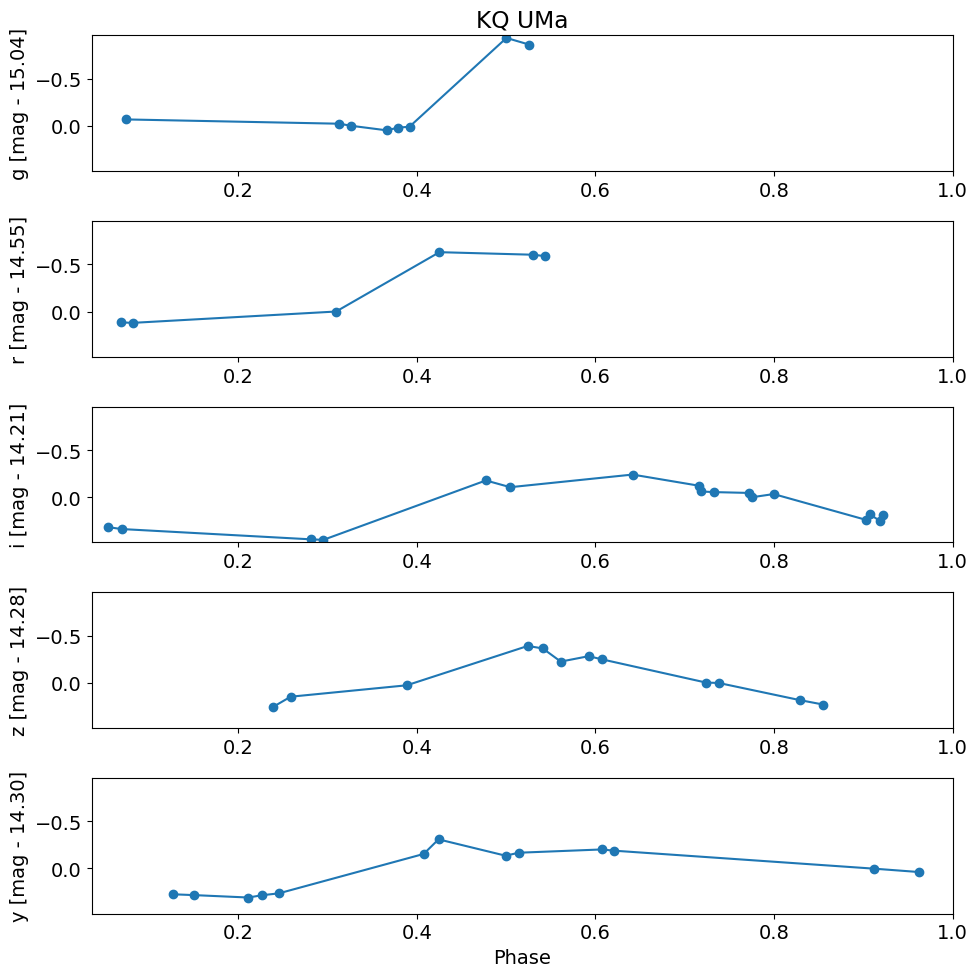
Plot versus the periodic phase instead of epoch#
Plot versus phase using known RR Lyr period from Simbad (table J/AJ/132/1202/table4).
period = 0.48636 # days, from Simbad
# convert flux in Jy to magnitudes
t = (detection_TAP_results['obsTime'] % period) / period
mag = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
magmean = 0.0*mag
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
magmean[detection_TAP_results['filterType'] == filter] = TAP_results[f'{filter}meanpsfmag'][0]
dmag = mag - magmean
dmag1 = dmag[detection_TAP_results['psfQfPerfect'] > 0.9]
# fix the x and y axis ranges
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
# flip axis direction for magnitude
ylim = np.array([dmag1.max(), dmag1.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where((detection_TAP_results['filterType'] == filter) & (detection_TAP_results['psfQfPerfect'] > 0.9))[0]
w = w[np.argsort(t[w])]
pylab.plot(t[w], dmag[w], '-o')
pylab.ylabel('{} [mag - {:.2f}]'.format(filter, magmean[w[0]]))
pylab.xlim(xlim)
pylab.ylim(ylim)
if i == 0:
pylab.title(objname)
pylab.xlabel('Phase')
pylab.tight_layout()
Repeat search using eclipsing binary KIC 2161623#
From Villanova Kepler Eclipsing Binaries
objname = 'KIC 2161623'
coords = Mast.resolve_object(objname)
ra, dec = coords.ra.value, coords.dec.value
radius = 1.0/3600.0 # radius = 1 arcsec
query = """
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag, rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',{},{},{}))=1
AND nDetections > 1
""".format(ra, dec, radius)
print(query)
job = TAP_service.run_async(query)
TAP_results = job.to_table()
TAP_results
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag, rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',291.744461,37.59102,0.0002777777777777778))=1
AND nDetections > 1
| objid | ramean | decmean | ndetections | ng | nr | ni | nz | ny | gmeanpsfmag | rmeanpsfmag | imeanpsfmag | zmeanpsfmag | ymeanpsfmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg | deg | mag | mag | mag | mag | mag | |||||||
| int64 | float64 | float64 | int16 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 |
| 153102917444859851 | 291.74446283634614 | 37.59099888154969 | 67 | 10 | 16 | 12 | 15 | 14 | 14.5998 | 14.2821 | 14.1587 | 14.2004 | 14.0672 |
Get Repeated Detection Information#
This time include the psfQfPerfect limit directly in the database query.
objid = TAP_results['objid'][0]
query = """
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID={}
AND psfQfPerfect >= 0.9
ORDER BY filterID, obsTime
""".format(objid)
print(query)
job = TAP_service.run_async(query)
detection_TAP_results = job.to_table()
# add magnitude and difference from mean
detection_TAP_results['magmean'] = 0.0
for i, filter in enumerate([b'g', b'r', b'i', b'z', b'y']):
detection_TAP_results['magmean'][detection_TAP_results['filterType'] == filter] = TAP_results[filter.decode('ascii')+'meanpsfmag'][0]
detection_TAP_results['mag'] = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
detection_TAP_results['dmag'] = detection_TAP_results['mag']-detection_TAP_results['magmean']
detection_TAP_results
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID=153102917444859851
AND psfQfPerfect >= 0.9
ORDER BY filterID, obsTime
| objID | detectID | filterid | filterType | obsTime | ra | dec | psfFlux | psfFluxErr | psfMajorFWHM | psfMinorFWHM | psfQfPerfect | apFlux | apFluxErr | infoFlag | infoFlag2 | infoFlag3 | magmean | mag | dmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d | deg | deg | arcsec | arcsec | |||||||||||||||
| int64 | int64 | uint8 | object | float64 | float64 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int64 | int32 | int32 | float64 | float64 | float64 |
| 153102917444859851 | 90150443710000088 | 1 | g | 55002.5047803 | 291.74446391 | 37.5910006 | 0.00501008 | 6.65687e-06 | 1.98744 | 1.81211 | 0.999421 | 0.00517413 | 7.31225e-06 | 102760517 | 128 | 124815424 | 0.0 | 14.650388503074646 | 14.650388503074646 |
| 153102917444859851 | 90151959710000087 | 1 | g | 55002.5199475 | 291.74446611 | 37.59100156 | 0.00487334 | 6.48844e-06 | 1.98487 | 1.70793 | 0.999234 | 0.00511542 | 7.27714e-06 | 102760517 | 128 | 124815424 | 0.0 | 14.680433416366578 | 14.680433416366578 |
| 153102917444859851 | 131534996560000129 | 1 | g | 55416.3502179 | 291.74446082 | 37.59099692 | 0.00525622 | 8.41646e-06 | 1.62403 | 1.57893 | 0.998566 | 0.0052992 | 8.62578e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.598316097259522 | 14.598316097259522 |
| 153102917444859851 | 131536180650000138 | 1 | g | 55416.362065 | 291.74445937 | 37.59099733 | 0.00533028 | 8.62169e-06 | 1.32081 | 1.27186 | 0.998757 | 0.00540082 | 8.91577e-06 | 102760517 | 128 | 124815424 | 0.0 | 14.583125257492066 | 14.583125257492066 |
| 153102917444859851 | 204528930560000076 | 1 | g | 56146.2895546 | 291.74445914 | 37.59098073 | 0.00522399 | 8.43794e-06 | 1.33369 | 1.29211 | 0.999758 | 0.00532506 | 8.88465e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.604994201660157 | 14.604994201660157 |
| 153102917444859851 | 204530041560000085 | 1 | g | 56146.3006673 | 291.74446486 | 37.59097649 | 0.00523539 | 8.44281e-06 | 1.42883 | 1.22683 | 0.964646 | 0.00532419 | 8.85594e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.602627301216126 | 14.602627301216126 |
| 153102917444859851 | 241032265120000094 | 1 | g | 56511.322909 | 291.74445657 | 37.59099752 | 0.00528641 | 8.3427e-06 | 1.15123 | 1.10789 | 0.998722 | 0.00537484 | 8.8135e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.592098140716553 | 14.592098140716553 |
| 153102917444859851 | 241033321120000081 | 1 | g | 56511.3334693 | 291.74445945 | 37.59099643 | 0.00527889 | 8.31263e-06 | 1.07898 | 0.987323 | 0.995279 | 0.0053346 | 8.77771e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.593643689155579 | 14.593643689155579 |
| 153102917444859851 | 91252543710000102 | 2 | r | 55013.5256677 | 291.74445985 | 37.59099454 | 0.00700243 | 8.7848e-06 | 1.90187 | 1.68689 | 0.998962 | 0.00723513 | 9.82915e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.28687825202942 | 14.28687825202942 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 153102917444859851 | 160460087410000136 | 5 | y | 55705.6010605 | 291.74446372 | 37.59099667 | 0.00838914 | 2.4518e-05 | 0.892294 | 0.785714 | 0.997498 | 0.00850705 | 2.38093e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.090706253051758 | 14.090706253051758 |
| 153102917444859851 | 160461274410000134 | 5 | y | 55705.6129264 | 291.74446479 | 37.59099484 | 0.00846376 | 2.45964e-05 | 0.843567 | 0.719354 | 0.982199 | 0.00845989 | 2.36938e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.081092023849488 | 14.081092023849488 |
| 153102917444859851 | 171734372010000127 | 5 | y | 55818.3439014 | 291.74446419 | 37.59099973 | 0.00899537 | 2.60801e-05 | 1.25605 | 0.963624 | 0.997509 | 0.00893125 | 2.37426e-05 | 102760517 | 128 | 57706560 | 0.0 | 14.014952325820923 | 14.014952325820923 |
| 153102917444859851 | 171735523010000129 | 5 | y | 55818.3554064 | 291.74446058 | 37.59100208 | 0.00893885 | 2.61524e-05 | 1.2684 | 0.941259 | 0.997929 | 0.00898301 | 2.38145e-05 | 102760517 | 128 | 40929344 | 0.0 | 14.021796131134034 | 14.021796131134034 |
| 153102917444859851 | 194462075640000160 | 5 | y | 56045.6209324 | 291.7444627 | 37.59099914 | 0.00901312 | 2.46438e-05 | 1.41157 | 1.29344 | 0.998375 | 0.00900578 | 2.28883e-05 | 102760517 | 128 | 57706560 | 0.0 | 14.012811923027039 | 14.012811923027039 |
| 153102917444859851 | 194462752640000158 | 5 | y | 56045.6277083 | 291.74445989 | 37.59099718 | 0.00881642 | 2.44413e-05 | 1.40377 | 1.19898 | 0.999512 | 0.00888709 | 2.26963e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.03676941394806 | 14.03676941394806 |
| 153102917444859851 | 213319324300000159 | 5 | y | 56234.1934155 | 291.74446373 | 37.5910009 | 0.00851151 | 2.42822e-05 | 1.04504 | 0.849606 | 0.99497 | 0.00842352 | 2.41261e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.074983739852906 | 14.074983739852906 |
| 153102917444859851 | 213319880300000163 | 5 | y | 56234.1989869 | 291.74446915 | 37.59100079 | 0.00852216 | 2.36733e-05 | 1.03871 | 0.843969 | 0.994977 | 0.00833225 | 2.42534e-05 | 102760517 | 128 | 74483776 | 0.0 | 14.073625946044922 | 14.073625946044922 |
| 153102917444859851 | 245335559360000173 | 5 | y | 56554.3560604 | 291.74446488 | 37.59099874 | 0.00870705 | 1.611e-05 | 1.42906 | 1.38935 | 0.959646 | 0.00886884 | 1.39697e-05 | 102760517 | 128 | 40929344 | 0.0 | 14.050322318077088 | 14.050322318077088 |
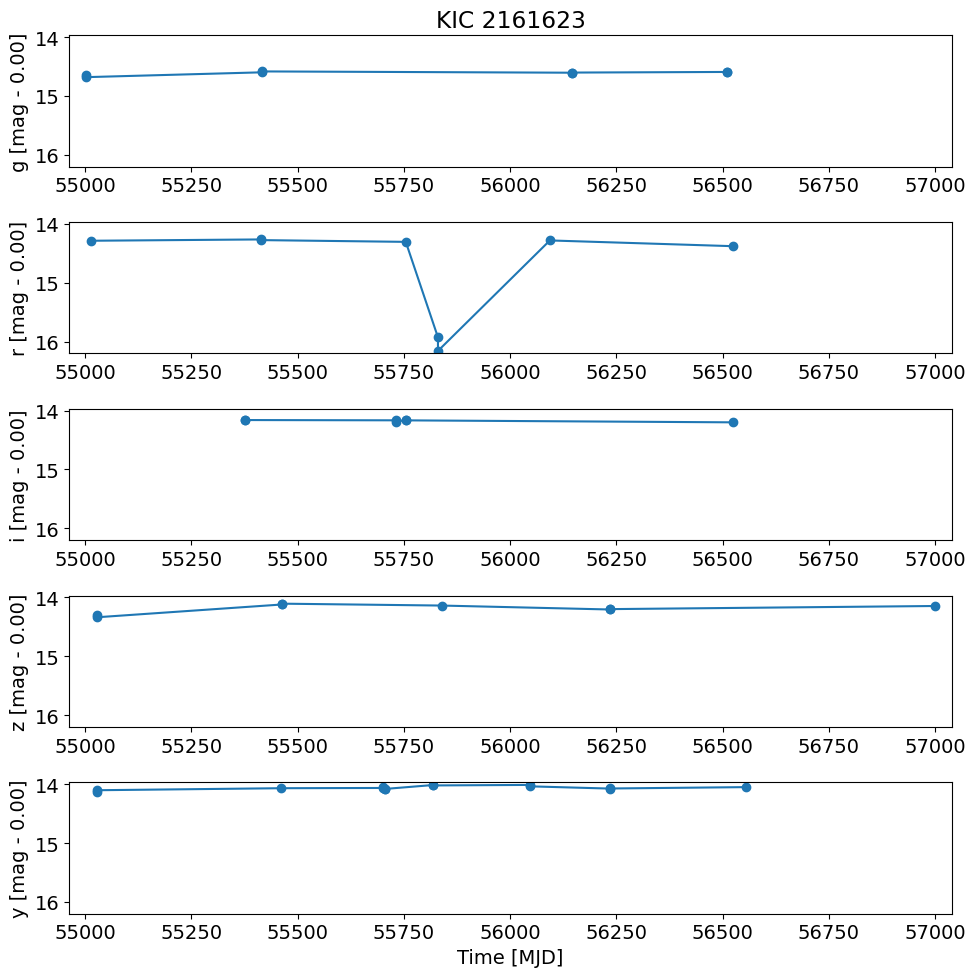
t = detection_TAP_results['obsTime']
dmag = detection_TAP_results['dmag']
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
ylim = np.array([dmag.max(), dmag.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where(detection_TAP_results['filterType'] == filter)[0]
pylab.plot(t[w], dmag[w], '-o')
magmean = detection_TAP_results['magmean'][w[0]]
pylab.ylabel('{} [mag - {:.2f}]'.format(filter, magmean))
pylab.xlim(xlim)
pylab.ylim(ylim)
if i == 0:
pylab.title(objname)
pylab.xlabel('Time [MJD]')
pylab.tight_layout()
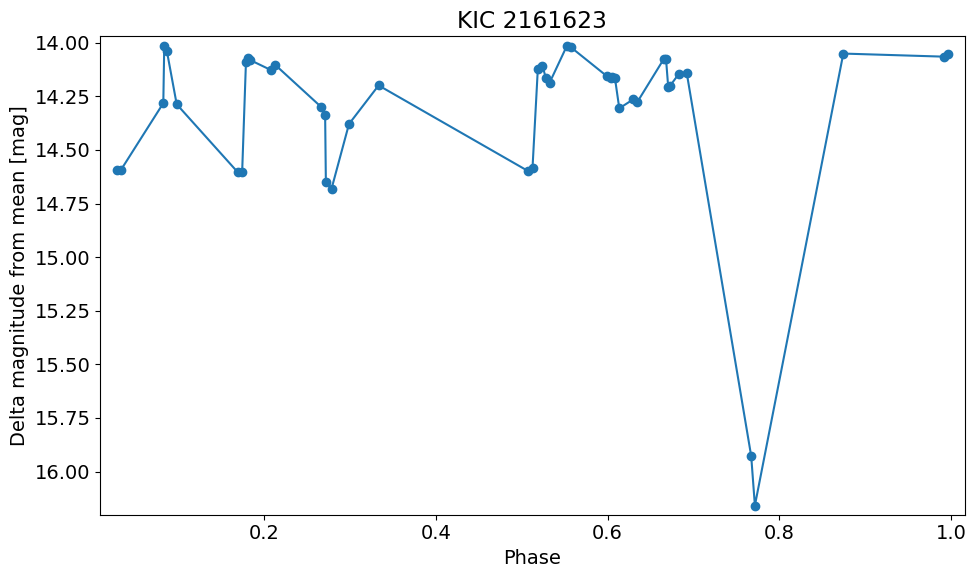
Plot versus phase using known period#
Eclipsing binaries basically vary by same amount in all filters since it is a geometrical effect, so combine the data into a single light curve. Wrap using known period and plot versus phase.
period = 2.2834698
bjd0 = 54999.599837
t = ((detection_TAP_results['obsTime']-bjd0) % period) / period
dmag = detection_TAP_results['dmag']
w = np.argsort(t)
t = t[w]
dmag = dmag[w]
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
ylim = np.array([dmag.max(), dmag.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 6))
pylab.plot(t, dmag, '-o')
pylab.xlim(xlim)
pylab.ylim(ylim)
pylab.xlabel('Phase')
pylab.ylabel('Delta magnitude from mean [mag]')
pylab.title(objname)
pylab.tight_layout()
Repeat search for another eclipsing binary KIC 8153568#
objname = 'KIC 8153568'
coords = Mast.resolve_object(objname)
ra, dec = coords.ra.value, coords.dec.value
radius = 1.0/3600.0 # radius = 1 arcsec
query = """
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag, rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',{},{},{}))=1
AND nDetections > 1
""".format(ra, dec, radius)
print(query)
job = TAP_service.run_async(query)
TAP_results = job.to_table()
TAP_results
SELECT objID, RAMean, DecMean, nDetections, ng, nr, ni, nz, ny, gMeanPSFMag, rMeanPSFMag, iMeanPSFMag, zMeanPSFMag, yMeanPSFMag
FROM dbo.MeanObjectView
WHERE
CONTAINS(POINT('ICRS', RAMean, DecMean),CIRCLE('ICRS',286.90445,44.00551,0.0002777777777777778))=1
AND nDetections > 1
| objid | ramean | decmean | ndetections | ng | nr | ni | nz | ny | gmeanpsfmag | rmeanpsfmag | imeanpsfmag | zmeanpsfmag | ymeanpsfmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg | deg | mag | mag | mag | mag | mag | |||||||
| int64 | float64 | float64 | int16 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 |
| 160802869044447231 | 286.90445005122217 | 44.00547945471509 | 88 | 16 | 15 | 31 | 10 | 16 | 15.1825 | 14.9899 | 14.8907 | 15.1999 | 14.8484 |
objid = TAP_results['objid'][0]
query = """
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID={}
AND psfQfPerfect >= 0.9
ORDER BY filterID, obsTime
""".format(objid)
print(query)
job = TAP_service.run_async(query)
detection_TAP_results = job.to_table()
# add magnitude and difference from mean
detection_TAP_results['magmean'] = 0.0
for i, filter in enumerate([b'g', b'r', b'i', b'z', b'y']):
detection_TAP_results['magmean'][detection_TAP_results['filterType'] == filter] = TAP_results[filter.decode('ascii')+'meanpsfmag'][0]
detection_TAP_results['mag'] = -2.5*np.log10(detection_TAP_results['psfFlux']) + 8.90
detection_TAP_results['dmag'] = detection_TAP_results['mag']-detection_TAP_results['magmean']
detection_TAP_results
SELECT
objID, detectID, Detection.filterID as filterID, Filter.filterType, obsTime, ra, dec,
psfFlux, psfFluxErr, psfMajorFWHM, psfMinorFWHM, psfQfPerfect,
apFlux, apFluxErr, infoFlag, infoFlag2, infoFlag3
FROM Detection
NATURAL JOIN Filter
WHERE objID=160802869044447231
AND psfQfPerfect >= 0.9
ORDER BY filterID, obsTime
| objID | detectID | filterid | filterType | obsTime | ra | dec | psfFlux | psfFluxErr | psfMajorFWHM | psfMinorFWHM | psfQfPerfect | apFlux | apFluxErr | infoFlag | infoFlag2 | infoFlag3 | magmean | mag | dmag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d | deg | deg | arcsec | arcsec | |||||||||||||||
| int64 | int64 | uint8 | object | float64 | float64 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int64 | int32 | int32 | float64 | float64 | float64 |
| 160802869044447231 | 91336429430000113 | 1 | g | 55014.3646518 | 286.90443374 | 44.00547575 | 0.003109 | 5.98065e-06 | 1.78 | 1.23811 | 0.998335 | 0.00312447 | 5.69291e-06 | 102760517 | 128 | 7374912 | 0.0 | 15.168448233604432 | 15.168448233604432 |
| 160802869044447231 | 91337480430000116 | 1 | g | 55014.3751566 | 286.90443894 | 44.00547502 | 0.00313056 | 6.00144e-06 | 1.66939 | 1.32773 | 0.998197 | 0.00315613 | 5.71358e-06 | 102760517 | 128 | 124815424 | 0.0 | 15.160945200920105 | 15.160945200920105 |
| 160802869044447231 | 126057565120000092 | 1 | g | 55361.57592 | 286.90445317 | 44.00547451 | 0.00309573 | 6.72312e-06 | 1.7498 | 1.62422 | 0.998826 | 0.00310045 | 6.69387e-06 | 102760517 | 128 | 7374912 | 0.0 | 15.173092031478882 | 15.173092031478882 |
| 160802869044447231 | 126058741130000111 | 1 | g | 55361.587678 | 286.90445157 | 44.00548065 | 0.00302815 | 6.57752e-06 | 1.62839 | 1.52579 | 0.998387 | 0.0031062 | 6.76425e-06 | 102760517 | 128 | 7374912 | 0.0 | 15.197056674957276 | 15.197056674957276 |
| 160802869044447231 | 128744126310000284 | 1 | g | 55388.4415151 | 286.9044502 | 44.00547891 | 0.000806462 | 3.51444e-06 | 1.10132 | 0.992501 | 0.997631 | 0.000810416 | 3.45294e-06 | 102760517 | 128 | 124815424 | 0.0 | 16.633539938926695 | 16.633539938926695 |
| 160802869044447231 | 128745341310000249 | 1 | g | 55388.4536712 | 286.90445043 | 44.00547936 | 0.000981762 | 3.82737e-06 | 1.1085 | 1.07441 | 0.997491 | 0.000985584 | 3.78859e-06 | 102760517 | 128 | 124815424 | 0.0 | 16.41998424530029 | 16.41998424530029 |
| 160802869044447231 | 164544841720000097 | 1 | g | 55746.4486739 | 286.90445315 | 44.00547646 | 0.00306655 | 6.32653e-06 | 1.32851 | 1.16693 | 0.99831 | 0.00304733 | 6.63894e-06 | 102760517 | 128 | 40929344 | 0.0 | 15.183375024795533 | 15.183375024795533 |
| 160802869044447231 | 164744408530000111 | 1 | g | 55748.4443394 | 286.90445207 | 44.00547835 | 0.00304057 | 6.5461e-06 | 1.37153 | 1.23193 | 0.998544 | 0.00307828 | 6.69725e-06 | 102760517 | 128 | 7374912 | 0.0 | 15.192612552642823 | 15.192612552642823 |
| 160802869044447231 | 164745526530000113 | 1 | g | 55748.4555151 | 286.90445275 | 44.0054797 | 0.00304728 | 6.57104e-06 | 1.27531 | 1.26424 | 0.998519 | 0.00308564 | 6.71944e-06 | 102760517 | 128 | 7374912 | 0.0 | 15.190218830108643 | 15.190218830108643 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 160802869044447231 | 136027009120000145 | 5 | y | 55461.2702758 | 286.90445503 | 44.00547827 | 0.00429769 | 1.96242e-05 | 1.48652 | 1.38968 | 0.998686 | 0.00423395 | 1.58905e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.81691255569458 | 14.81691255569458 |
| 160802869044447231 | 136028204120000148 | 5 | y | 55461.2822322 | 286.90445994 | 44.00548229 | 0.00417461 | 2.0278e-05 | 1.76506 | 1.713 | 0.998705 | 0.00419736 | 1.56164e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.8484601020813 | 14.8484601020813 |
| 160802869044447231 | 159959993400000147 | 5 | y | 55700.6001108 | 286.90444692 | 44.00548086 | 0.00400983 | 1.95314e-05 | 1.04048 | 0.898651 | 0.998029 | 0.00396177 | 1.71112e-05 | 102760517 | 128 | 40929344 | 0.0 | 14.89218487739563 | 14.89218487739563 |
| 160802869044447231 | 159961165400000142 | 5 | y | 55700.611833 | 286.90444463 | 44.00548094 | 0.00409667 | 1.92559e-05 | 0.876026 | 0.741052 | 0.998806 | 0.00403949 | 1.75641e-05 | 102760517 | 128 | 7374912 | 0.0 | 14.86892237663269 | 14.86892237663269 |
| 160802869044447231 | 160459626400000159 | 5 | y | 55705.5964454 | 286.9044489 | 44.0054811 | 0.00331343 | 1.64053e-05 | 1.0118 | 0.84848 | 0.997465 | 0.00327993 | 1.48499e-05 | 102760517 | 128 | 124815424 | 0.0 | 15.099305653572083 | 15.099305653572083 |
| 160802869044447231 | 160460795400000171 | 5 | y | 55705.6081345 | 286.90444684 | 44.00547964 | 0.00321194 | 1.64645e-05 | 1.08121 | 0.990937 | 0.997873 | 0.0031801 | 1.44718e-05 | 102760517 | 128 | 124815424 | 0.0 | 15.133081221580506 | 15.133081221580506 |
| 160802869044447231 | 171023586640000136 | 5 | y | 55811.2360427 | 286.9044525 | 44.00548357 | 0.0042144 | 1.82169e-05 | 0.723151 | 0.662475 | 0.9978 | 0.00425246 | 1.73593e-05 | 102760517 | 128 | 7342144 | 0.0 | 14.838160419464112 | 14.838160419464112 |
| 160802869044447231 | 171024776640000151 | 5 | y | 55811.2479472 | 286.90445276 | 44.00548227 | 0.00425027 | 1.862e-05 | 0.763658 | 0.692114 | 0.997703 | 0.00431336 | 1.74107e-05 | 102760517 | 128 | 7342144 | 0.0 | 14.828958654403687 | 14.828958654403687 |
| 160802869044447231 | 245335002470000153 | 5 | y | 56554.3504901 | 286.90445085 | 44.00547784 | 0.00427093 | 1.24367e-05 | 1.46348 | 1.40394 | 0.972607 | 0.00426433 | 9.76066e-06 | 102760517 | 128 | 7374912 | 0.0 | 14.823693776130677 | 14.823693776130677 |
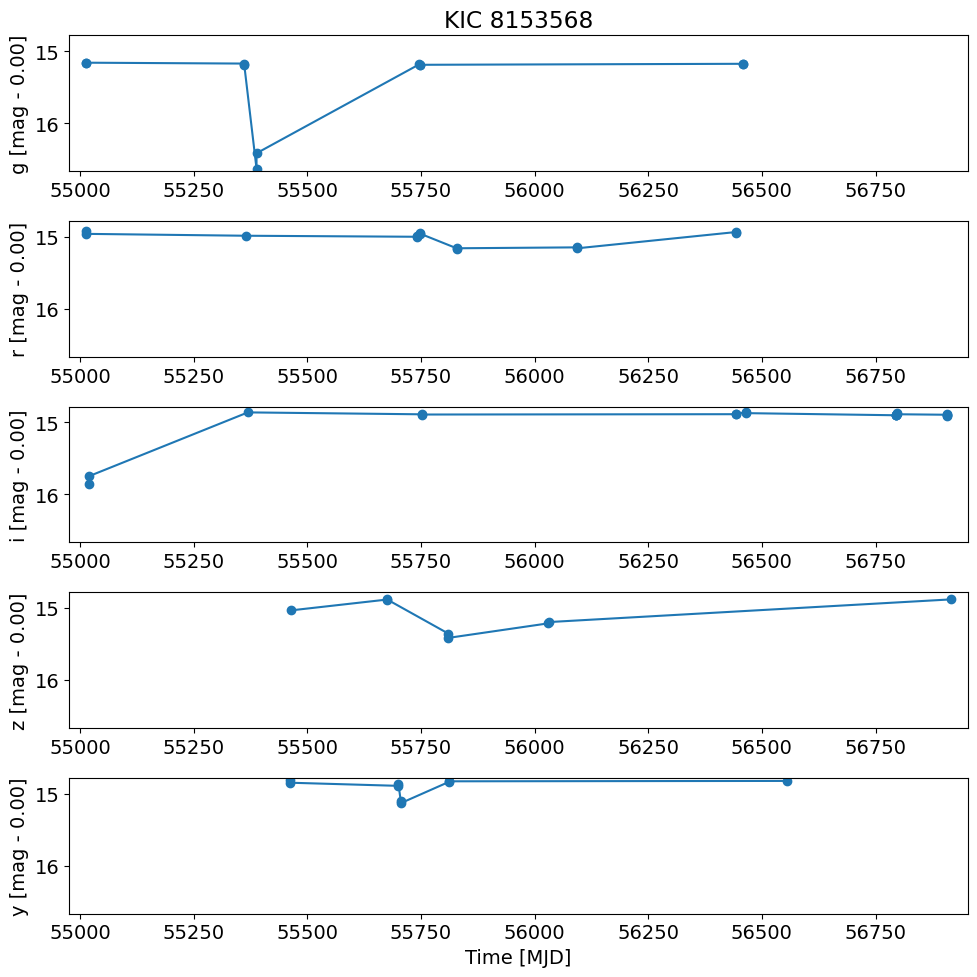
t = detection_TAP_results['obsTime']
dmag = detection_TAP_results['dmag']
xlim = np.array([t.min(), t.max()])
xlim = xlim + np.array([-1, 1]) * 0.02 * (xlim[1] - xlim[0])
ylim = np.array([dmag.max(), dmag.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (10, 10))
for i, filter in enumerate(['g', 'r', 'i', 'z', 'y']):
pylab.subplot(511+i)
w = np.where(detection_TAP_results['filterType'] == filter)[0]
pylab.plot(t[w], dmag[w], '-o')
magmean = detection_TAP_results['magmean'][w[0]]
pylab.ylabel('{} [mag - {:.2f}]'.format(filter, magmean))
pylab.xlim(xlim)
pylab.ylim(ylim)
if i == 0:
pylab.title(objname)
pylab.xlabel('Time [MJD]')
pylab.tight_layout()
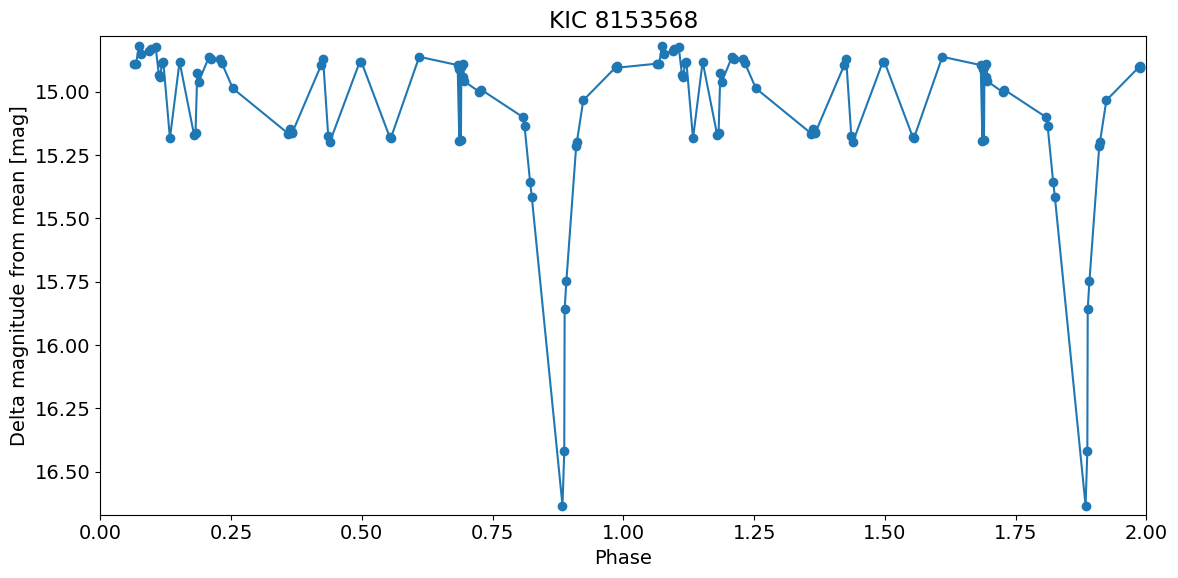
Eclipsing binaries basically vary by same amount in all filters since it is a geometrical effect, so combine the data into a single light curve.
Wrap using known period and plot versus phase. Plot two periods of the light curve this time.
This nice light curve appears to show a secondary eclipse.
period = 3.6071431
bjd0 = 54999.289794
t = ((detection_TAP_results['obsTime']-bjd0) % period) / period
dmag = detection_TAP_results['dmag']
w = np.argsort(t)
# extend to two periods
nw = len(w)
w = np.append(w, w)
t = t[w]
# add one to second period
t[-nw:] += 1
dmag = dmag[w]
xlim = [0, 2.0]
ylim = np.array([dmag.max(), dmag.min()])
ylim = ylim + np.array([-1, 1]) * 0.02 * (ylim[1] - ylim[0])
pylab.rcParams.update({'font.size': 14})
pylab.figure(1, (12, 6))
pylab.plot(t, dmag, '-o')
pylab.xlim(xlim)
pylab.ylim(ylim)
pylab.xlabel('Phase')
pylab.ylabel('Delta magnitude from mean [mag]')
pylab.title(objname)
pylab.tight_layout()
Additional Resources#
Table Access Protocol#
IVOA standard for RESTful web service access to tabular data
http://www.ivoa.net/documents/TAP/
PanSTARRS 1 DR 2#
Catalog for PanSTARRS with additional Detection information
https://outerspace.stsci.edu/display/PANSTARRS/
Astronomical Query Data Language (2.0)#
IVOA standard for querying astronomical data in tabular format, with geometric search support
http://www.ivoa.net/documents/latest/ADQL.html
PyVO#
an affiliated package for astropy
find and retrieve astronomical data available from archives that support standard IVOA virtual observatory service protocols.
https://pyvo.readthedocs.io/en/latest/index.html
About this Notebook#
Authors: Rick White & Theresa Dower, with edits from T. Dutkiewicz
For support, please contact the Archive HelpDesk at archive@stsci.edu.

