This notebook is now part of the JWST-pipeline-notebooks repository and will be removed from this repository by November 30, 2024. Please access the notebook at its new location.
Advanced: NIRISS Imaging Tutorial Notebook#
Table of Contents#
Introduction
Examining uncalibrated data products
Stage 1 Processing
Stage 2 Processing
Stage 3 Processing
Visualize Detected Sources
Date published: January 24, 2024
1. Introduction#
In this notebook, we will process a NIRISS imaging dataset through the JWST calibration pipeline. The example dataset is from Program ID 1475 (PI: Boyer, CoI: Volk) which is a sky flat calibration program. NIRCam is used as the primary instrument with NIRISS as a coordinated parallel instrument. The NIRISS imaging dataset uses a 17-step dither pattern.
For illustrative purposes, we focus on data taken through the NIRISS F150W filter and start with uncalibrated data products. The files are named
jw01475006001_02201_000nn_nis_uncal.fits, where nn refers to the dither step number which ranges from 01 - 17. See the “File Naming Schemes” documentation to learn more about the file naming convention.
In this notebook, we assume all uncalibrated fits files are saved in a directory named 1475_f150w.
Install pipeline and dependencies#
To make sure that the pipeline version is compatabile with the steps discussed below and the required dependencies and packages are installed, you can create a fresh conda environment and install the provided requirements.txt file:
conda create -n niriss_imaging_pipeline python=3.11
conda activate niriss_imaging_pipeline
pip install -r requirements.txt
Imports#
import os
import glob
import zipfile
import numpy as np
import urllib.request
from IPython.display import Image
# For visualizing images
import jdaviz
from jdaviz import Imviz
# Astropy routines for visualizing detected sources:
from astropy.table import Table
from astropy.coordinates import SkyCoord
# Configure CRDS
os.environ["CRDS_PATH"] = 'crds_cache'
os.environ["CRDS_SERVER_URL"] = "https://jwst-crds.stsci.edu"
# for JWST calibration pipeline
import jwst
from jwst import datamodels
from jwst.pipeline import Detector1Pipeline, Image2Pipeline, Image3Pipeline
from jwst.associations import asn_from_list
from jwst.associations.lib.rules_level3_base import DMS_Level3_Base
# To confirm which version of the pipeline you're running:
print(f"jwst pipeline version: {jwst.__version__}")
print(f"jdaviz version: {jdaviz.__version__}")
jwst pipeline version: 1.16.1
jdaviz version: 4.0.0
Download uncalibrated data products#
# APT program ID number:
pid = '01475'
# Set up directory to download uncalibrated data files:
data_dir = '1475_f150w/'
# Create directory if it does not exist
if not os.path.isdir(data_dir):
os.mkdir(data_dir)
# Download uncalibrated data from Box into current directory:
boxlink = 'https://data.science.stsci.edu/redirect/JWST/jwst-data_analysis_tools/niriss_imaging/1475_f150w.zip'
boxfile = os.path.join(data_dir, '1475_f150w.zip')
urllib.request.urlretrieve(boxlink, boxfile)
zf = zipfile.ZipFile(boxfile, 'r')
zf.extractall()
2. Examining uncalibrated data products#
uncal_files = sorted(glob.glob(os.path.join(data_dir, '*_uncal.fits')))
Look at the first file to determine exposure parameters and practice using JWST datamodels#
# print file name
print(uncal_files[0])
# Open file as JWST datamodel
examine = datamodels.open(uncal_files[0])
# Print out exposure info
print("Instrument: " + examine.meta.instrument.name)
print("Filter: " + examine.meta.instrument.filter)
print("Pupil: " + examine.meta.instrument.pupil)
print("Number of integrations: {}".format(examine.meta.exposure.nints))
print("Number of groups: {}".format(examine.meta.exposure.ngroups))
print("Readout pattern: " + examine.meta.exposure.readpatt)
print("Dither position number: {}".format(examine.meta.dither.position_number))
print("\n")
1475_f150w/jw01475006001_02201_00001_nis_uncal.fits
Instrument: NIRISS
Filter: CLEAR
Pupil: F150W
Number of integrations: 1
Number of groups: 16
Readout pattern: NIS
Dither position number: 1
From the above, we confirm that the data file is for the NIRISS instrument using the F150W filter in the Pupil Wheel crossed with the CLEAR filter in the Filter Wheel. This observation uses the NIS readout pattern, 16 groups per integration, and 1 integration per exposure. This data file is the 1st dither position in this exposure sequence. For more information about how JWST exposures are defined by up-the-ramp sampling, see the Understanding Exposure Times JDox article.
This metadata will be the same for all exposures in this observation other than the dither position number.
Display uncalibrated image#
We can visualize an uncalibrated dataset that will show detector artifacts that will be removed when calibrating the data through the DETECTOR1 stage of the pipeline. Uncalibrated data files thus are 4D: nintegrations x ngroups x nrows x ncols. Here, we are visualizing the full detector (i.e., all columns and rows) and the 1st group.
We are using the Imviz tool within the jdaviz package to visualize the NIRISS image.
# Create an Imviz instance and set up default viewer
imviz_uncal = Imviz()
viewer_uncal = imviz_uncal.default_viewer
# Read in the science array for our visualization dataset:
uncal_science = examine.data
# Load the dataset into Imviz
imviz_uncal.load_data(uncal_science[0, 0, :, :])
# Visualize the dataset:
imviz_uncal.show()

Adjust settings for the viewer:
plotopt = imviz_uncal.plugins['Plot Options']
plotopt.stretch_function = 'sqrt'
plotopt.image_colormap = 'Viridis'
plotopt.stretch_preset = '99.5%'
plotopt.zoom_radius = 1024
The viewer looks like this:
viewer_uncal.save('./uncal_science.png')
Image('./uncal_science.png')
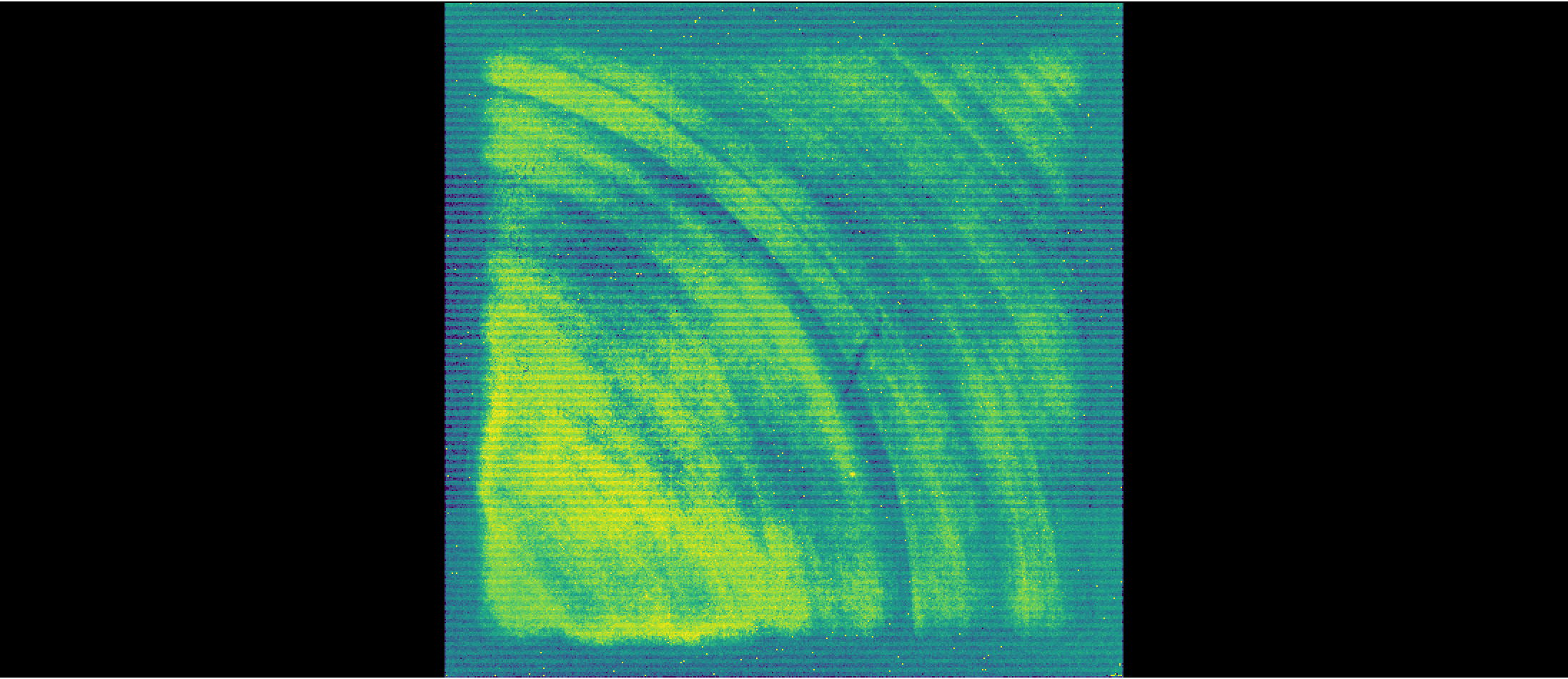
3. Stage 1 Processing #
Run the datasets through the Detector1 stage of the pipeline to apply detector level calibrations and create a countrate data product where slopes are fitted to the integration ramps. These *_rate.fits products are 2D (nrows x ncols), averaged over all integrations. 3D countrate data products (*_rateints.fits) are also created (nintegrations x nrows x ncols) which have the fitted ramp slopes for each integration.
The pipeline documentation discusses how to run the pipeline in the Python Interface, including how to configure pipeline steps and override reference files. By returning the results of the pipeline to a variable, the pipeline returns a datamodel. Note that the pipeline.call() method is preferred over the pipeline.run() method.
By default, all steps in the Detector1 stage of the pipeline are run for NIRISS except: the ipc correction step and the gain_scale step. Note that while the persistence step is set to run by default, this step does not automatically correct the science data for persistence. The persistence step creates a *_trapsfilled.fits file which is a model that records the number of traps filled at each pixel at the end of an exposure. This file would be used as an input to the persistence step, via the input_trapsfilled argument, to correct a science exposure for persistence. Since persistence is not well calibrated for NIRISS, we do not perform a persistence correction and thus turn off this step to speed up calibration and to not create files that will not be used in the subsequent analysis. This step can be turned off when running the pipeline in Python by doing:
rate_result = Detector1Pipeline.call(uncal,
steps={'persistence': {'skip': True}})
The charge_migration step is particularly important for NIRISS images to mitigate apparent flux loss in resampled images due to the spilling of charge from a central pixel into its neighboring pixels (see Goudfrooij et al. 2023 for details). Charge migration occurs when the accumulated charge in a central pixel exceeds a certain signal limit, which is ~25,000 ADU. This step is turned on by default for NIRISS imaging, Wide Field Slitless Spectroscopy (WFSS), and Aperture Masking Interferometry (AMI) modes when using CRDS contexts of jwst_1159.pmap or later. Different signal limits for each filter are provided by the pars-chargemigrationstep parameter files. Users can specify a different signal limit by running this step with the signal_threshold flag and entering another signal limit in units of ADU.
As of CRDS context jwst_1155.pmap and later, the jump step of the DETECTOR1 stage of the pipeline will remove residuals associated with snowballs for NIRISS imaging, WFSS, and AMI modes. The default parameters for this correction, where expand_large_events set to True turns on the snowball halo removal algorithm, are specified in the pars-jumpstep parameter reference files. Users may wish to alter parameters to optimize removal of snowball residuals. Available parameters are discussed in the Detection and Flagging of Showers and Snowballs in JWST Technical Report (Regan 2023).
# Define directory to save output from detector1
det1_dir = 'detector1/'
# Create directory if it does not exist
if not os.path.isdir(det1_dir):
os.mkdir(det1_dir)
# Run Detector1 stage of pipeline, specifying:
# output directory to save *_rate.fits files
# save_results flag set to True so the rate files are saved
# skipping the persistence step
for uncal in uncal_files:
rate_result = Detector1Pipeline.call(uncal,
output_dir=det1_dir,
save_results=True,
steps={'persistence': {'skip': True}})
2024-12-10 20:13:44,678 - CRDS - INFO - Calibration SW Found: jwst 1.16.1 (/opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst-1.16.1.dist-info)
2024-12-10 20:13:45,072 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_system_datalvl_0002.rmap 694 bytes (1 / 200 files) (0 / 717.9 K bytes)
2024-12-10 20:13:45,242 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_system_calver_0044.rmap 5.0 K bytes (2 / 200 files) (694 / 717.9 K bytes)
2024-12-10 20:13:45,369 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_system_0043.imap 385 bytes (3 / 200 files) (5.7 K / 717.9 K bytes)
2024-12-10 20:13:45,528 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_wavelengthrange_0024.rmap 1.4 K bytes (4 / 200 files) (6.1 K / 717.9 K bytes)
2024-12-10 20:13:45,724 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_wavecorr_0005.rmap 884 bytes (5 / 200 files) (7.5 K / 717.9 K bytes)
2024-12-10 20:13:45,877 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_superbias_0074.rmap 33.8 K bytes (6 / 200 files) (8.3 K / 717.9 K bytes)
2024-12-10 20:13:46,014 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_sflat_0026.rmap 20.6 K bytes (7 / 200 files) (42.1 K / 717.9 K bytes)
2024-12-10 20:13:46,150 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_saturation_0018.rmap 2.0 K bytes (8 / 200 files) (62.7 K / 717.9 K bytes)
2024-12-10 20:13:46,302 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_refpix_0015.rmap 1.6 K bytes (9 / 200 files) (64.7 K / 717.9 K bytes)
2024-12-10 20:13:46,504 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_readnoise_0025.rmap 2.6 K bytes (10 / 200 files) (66.3 K / 717.9 K bytes)
2024-12-10 20:13:46,642 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_photom_0013.rmap 958 bytes (11 / 200 files) (68.9 K / 717.9 K bytes)
2024-12-10 20:13:46,806 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pathloss_0007.rmap 1.1 K bytes (12 / 200 files) (69.8 K / 717.9 K bytes)
2024-12-10 20:13:46,922 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-whitelightstep_0001.rmap 777 bytes (13 / 200 files) (70.9 K / 717.9 K bytes)
2024-12-10 20:13:47,065 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-spec2pipeline_0013.rmap 2.1 K bytes (14 / 200 files) (71.7 K / 717.9 K bytes)
2024-12-10 20:13:47,252 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-resamplespecstep_0002.rmap 709 bytes (15 / 200 files) (73.8 K / 717.9 K bytes)
2024-12-10 20:13:47,367 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-outlierdetectionstep_0003.rmap 1.1 K bytes (16 / 200 files) (74.6 K / 717.9 K bytes)
2024-12-10 20:13:47,481 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-jumpstep_0005.rmap 810 bytes (17 / 200 files) (75.7 K / 717.9 K bytes)
2024-12-10 20:13:47,623 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-image2pipeline_0008.rmap 1.0 K bytes (18 / 200 files) (76.5 K / 717.9 K bytes)
2024-12-10 20:13:47,736 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-detector1pipeline_0003.rmap 1.1 K bytes (19 / 200 files) (77.5 K / 717.9 K bytes)
2024-12-10 20:13:47,851 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-darkpipeline_0003.rmap 872 bytes (20 / 200 files) (78.6 K / 717.9 K bytes)
2024-12-10 20:13:47,959 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_pars-darkcurrentstep_0001.rmap 622 bytes (21 / 200 files) (79.4 K / 717.9 K bytes)
2024-12-10 20:13:48,076 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_ote_0030.rmap 1.3 K bytes (22 / 200 files) (80.1 K / 717.9 K bytes)
2024-12-10 20:13:48,190 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_msaoper_0014.rmap 1.4 K bytes (23 / 200 files) (81.3 K / 717.9 K bytes)
2024-12-10 20:13:48,325 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_msa_0027.rmap 1.3 K bytes (24 / 200 files) (82.7 K / 717.9 K bytes)
2024-12-10 20:13:48,442 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_mask_0039.rmap 2.7 K bytes (25 / 200 files) (84.0 K / 717.9 K bytes)
2024-12-10 20:13:48,555 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_linearity_0017.rmap 1.6 K bytes (26 / 200 files) (86.7 K / 717.9 K bytes)
2024-12-10 20:13:48,669 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_ipc_0006.rmap 876 bytes (27 / 200 files) (88.2 K / 717.9 K bytes)
2024-12-10 20:13:48,783 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_ifuslicer_0017.rmap 1.5 K bytes (28 / 200 files) (89.1 K / 717.9 K bytes)
2024-12-10 20:13:48,893 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_ifupost_0019.rmap 1.5 K bytes (29 / 200 files) (90.7 K / 717.9 K bytes)
2024-12-10 20:13:49,008 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_ifufore_0017.rmap 1.5 K bytes (30 / 200 files) (92.2 K / 717.9 K bytes)
2024-12-10 20:13:49,125 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_gain_0023.rmap 1.8 K bytes (31 / 200 files) (93.7 K / 717.9 K bytes)
2024-12-10 20:13:49,244 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_fpa_0028.rmap 1.3 K bytes (32 / 200 files) (95.4 K / 717.9 K bytes)
2024-12-10 20:13:49,353 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_fore_0026.rmap 5.0 K bytes (33 / 200 files) (96.7 K / 717.9 K bytes)
2024-12-10 20:13:49,465 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_flat_0015.rmap 3.8 K bytes (34 / 200 files) (101.6 K / 717.9 K bytes)
2024-12-10 20:13:49,580 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_fflat_0026.rmap 7.2 K bytes (35 / 200 files) (105.5 K / 717.9 K bytes)
2024-12-10 20:13:49,688 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_extract1d_0018.rmap 2.3 K bytes (36 / 200 files) (112.7 K / 717.9 K bytes)
2024-12-10 20:13:49,799 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_disperser_0028.rmap 5.7 K bytes (37 / 200 files) (115.0 K / 717.9 K bytes)
2024-12-10 20:13:49,918 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_dflat_0007.rmap 1.1 K bytes (38 / 200 files) (120.7 K / 717.9 K bytes)
2024-12-10 20:13:50,029 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_dark_0069.rmap 32.6 K bytes (39 / 200 files) (121.8 K / 717.9 K bytes)
2024-12-10 20:13:50,198 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_cubepar_0015.rmap 966 bytes (40 / 200 files) (154.4 K / 717.9 K bytes)
2024-12-10 20:13:50,347 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_collimator_0026.rmap 1.3 K bytes (41 / 200 files) (155.3 K / 717.9 K bytes)
2024-12-10 20:13:50,481 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_camera_0026.rmap 1.3 K bytes (42 / 200 files) (156.7 K / 717.9 K bytes)
2024-12-10 20:13:50,661 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_barshadow_0007.rmap 1.8 K bytes (43 / 200 files) (158.0 K / 717.9 K bytes)
2024-12-10 20:13:50,824 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_area_0018.rmap 6.3 K bytes (44 / 200 files) (159.8 K / 717.9 K bytes)
2024-12-10 20:13:50,949 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_apcorr_0009.rmap 5.6 K bytes (45 / 200 files) (166.0 K / 717.9 K bytes)
2024-12-10 20:13:51,095 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nirspec_0383.imap 5.6 K bytes (46 / 200 files) (171.6 K / 717.9 K bytes)
2024-12-10 20:13:51,216 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_wfssbkg_0007.rmap 3.1 K bytes (47 / 200 files) (177.2 K / 717.9 K bytes)
2024-12-10 20:13:51,327 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_wavemap_0008.rmap 2.2 K bytes (48 / 200 files) (180.3 K / 717.9 K bytes)
2024-12-10 20:13:51,440 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_wavelengthrange_0006.rmap 862 bytes (49 / 200 files) (182.6 K / 717.9 K bytes)
2024-12-10 20:13:51,554 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_trappars_0004.rmap 753 bytes (50 / 200 files) (183.4 K / 717.9 K bytes)
2024-12-10 20:13:51,664 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_trapdensity_0005.rmap 705 bytes (51 / 200 files) (184.2 K / 717.9 K bytes)
2024-12-10 20:13:51,773 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_throughput_0005.rmap 1.3 K bytes (52 / 200 files) (184.9 K / 717.9 K bytes)
2024-12-10 20:13:51,893 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_superbias_0028.rmap 6.5 K bytes (53 / 200 files) (186.1 K / 717.9 K bytes)
2024-12-10 20:13:52,065 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_specwcs_0014.rmap 3.1 K bytes (54 / 200 files) (192.6 K / 717.9 K bytes)
2024-12-10 20:13:52,175 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_spectrace_0008.rmap 2.3 K bytes (55 / 200 files) (195.8 K / 717.9 K bytes)
2024-12-10 20:13:52,333 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_specprofile_0008.rmap 2.4 K bytes (56 / 200 files) (198.1 K / 717.9 K bytes)
2024-12-10 20:13:52,501 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_speckernel_0006.rmap 1.0 K bytes (57 / 200 files) (200.4 K / 717.9 K bytes)
2024-12-10 20:13:52,692 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_saturation_0015.rmap 829 bytes (58 / 200 files) (201.5 K / 717.9 K bytes)
2024-12-10 20:13:52,814 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_readnoise_0011.rmap 987 bytes (59 / 200 files) (202.3 K / 717.9 K bytes)
2024-12-10 20:13:52,937 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_photom_0035.rmap 1.3 K bytes (60 / 200 files) (203.3 K / 717.9 K bytes)
2024-12-10 20:13:53,114 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_persat_0007.rmap 674 bytes (61 / 200 files) (204.5 K / 717.9 K bytes)
2024-12-10 20:13:53,281 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pathloss_0003.rmap 758 bytes (62 / 200 files) (205.2 K / 717.9 K bytes)
2024-12-10 20:13:53,396 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pastasoss_0004.rmap 818 bytes (63 / 200 files) (206.0 K / 717.9 K bytes)
2024-12-10 20:13:53,519 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-undersamplecorrectionstep_0001.rmap 904 bytes (64 / 200 files) (206.8 K / 717.9 K bytes)
2024-12-10 20:13:53,639 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-tweakregstep_0012.rmap 3.1 K bytes (65 / 200 files) (207.7 K / 717.9 K bytes)
2024-12-10 20:13:53,753 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-spec2pipeline_0008.rmap 984 bytes (66 / 200 files) (210.8 K / 717.9 K bytes)
2024-12-10 20:13:53,866 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-sourcecatalogstep_0002.rmap 2.3 K bytes (67 / 200 files) (211.8 K / 717.9 K bytes)
2024-12-10 20:13:54,005 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-resamplestep_0002.rmap 687 bytes (68 / 200 files) (214.1 K / 717.9 K bytes)
2024-12-10 20:13:54,153 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-outlierdetectionstep_0004.rmap 2.7 K bytes (69 / 200 files) (214.8 K / 717.9 K bytes)
2024-12-10 20:13:54,324 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-jumpstep_0007.rmap 6.4 K bytes (70 / 200 files) (217.5 K / 717.9 K bytes)
2024-12-10 20:13:54,671 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-image2pipeline_0005.rmap 1.0 K bytes (71 / 200 files) (223.8 K / 717.9 K bytes)
2024-12-10 20:13:55,018 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-detector1pipeline_0002.rmap 1.0 K bytes (72 / 200 files) (224.9 K / 717.9 K bytes)
2024-12-10 20:13:55,270 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-darkpipeline_0002.rmap 868 bytes (73 / 200 files) (225.9 K / 717.9 K bytes)
2024-12-10 20:13:55,547 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-darkcurrentstep_0001.rmap 591 bytes (74 / 200 files) (226.8 K / 717.9 K bytes)
2024-12-10 20:13:55,739 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_pars-chargemigrationstep_0004.rmap 5.7 K bytes (75 / 200 files) (227.4 K / 717.9 K bytes)
2024-12-10 20:13:55,865 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_nrm_0002.rmap 663 bytes (76 / 200 files) (233.0 K / 717.9 K bytes)
2024-12-10 20:13:56,281 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_mask_0020.rmap 859 bytes (77 / 200 files) (233.7 K / 717.9 K bytes)
2024-12-10 20:13:56,639 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_linearity_0022.rmap 961 bytes (78 / 200 files) (234.5 K / 717.9 K bytes)
2024-12-10 20:13:56,854 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_ipc_0007.rmap 651 bytes (79 / 200 files) (235.5 K / 717.9 K bytes)
2024-12-10 20:13:57,201 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_gain_0011.rmap 797 bytes (80 / 200 files) (236.2 K / 717.9 K bytes)
2024-12-10 20:13:57,336 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_flat_0023.rmap 5.9 K bytes (81 / 200 files) (236.9 K / 717.9 K bytes)
2024-12-10 20:13:57,477 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_filteroffset_0010.rmap 853 bytes (82 / 200 files) (242.8 K / 717.9 K bytes)
2024-12-10 20:13:57,614 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_extract1d_0007.rmap 905 bytes (83 / 200 files) (243.7 K / 717.9 K bytes)
2024-12-10 20:13:57,765 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_drizpars_0004.rmap 519 bytes (84 / 200 files) (244.6 K / 717.9 K bytes)
2024-12-10 20:13:57,978 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_distortion_0025.rmap 3.4 K bytes (85 / 200 files) (245.1 K / 717.9 K bytes)
2024-12-10 20:13:58,212 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_dark_0031.rmap 6.8 K bytes (86 / 200 files) (248.5 K / 717.9 K bytes)
2024-12-10 20:13:58,333 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_area_0014.rmap 2.7 K bytes (87 / 200 files) (255.3 K / 717.9 K bytes)
2024-12-10 20:13:58,483 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_apcorr_0010.rmap 4.3 K bytes (88 / 200 files) (258.0 K / 717.9 K bytes)
2024-12-10 20:13:58,706 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_abvegaoffset_0004.rmap 1.4 K bytes (89 / 200 files) (262.3 K / 717.9 K bytes)
2024-12-10 20:13:58,945 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_niriss_0261.imap 5.7 K bytes (90 / 200 files) (263.6 K / 717.9 K bytes)
2024-12-10 20:13:59,126 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_wfssbkg_0004.rmap 7.2 K bytes (91 / 200 files) (269.3 K / 717.9 K bytes)
2024-12-10 20:13:59,295 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_wavelengthrange_0010.rmap 996 bytes (92 / 200 files) (276.5 K / 717.9 K bytes)
2024-12-10 20:13:59,421 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_tsophot_0003.rmap 896 bytes (93 / 200 files) (277.5 K / 717.9 K bytes)
2024-12-10 20:13:59,529 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_trappars_0003.rmap 1.6 K bytes (94 / 200 files) (278.4 K / 717.9 K bytes)
2024-12-10 20:13:59,678 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_trapdensity_0003.rmap 1.6 K bytes (95 / 200 files) (280.0 K / 717.9 K bytes)
2024-12-10 20:13:59,796 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_superbias_0018.rmap 16.2 K bytes (96 / 200 files) (281.7 K / 717.9 K bytes)
2024-12-10 20:14:00,013 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_specwcs_0022.rmap 7.1 K bytes (97 / 200 files) (297.8 K / 717.9 K bytes)
2024-12-10 20:14:00,161 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_saturation_0010.rmap 2.2 K bytes (98 / 200 files) (305.0 K / 717.9 K bytes)
2024-12-10 20:14:00,273 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_readnoise_0025.rmap 23.2 K bytes (99 / 200 files) (307.1 K / 717.9 K bytes)
2024-12-10 20:14:00,440 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_psfmask_0008.rmap 28.4 K bytes (100 / 200 files) (330.3 K / 717.9 K bytes)
2024-12-10 20:14:00,596 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_photom_0028.rmap 3.4 K bytes (101 / 200 files) (358.7 K / 717.9 K bytes)
2024-12-10 20:14:00,706 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_persat_0005.rmap 1.6 K bytes (102 / 200 files) (362.0 K / 717.9 K bytes)
2024-12-10 20:14:00,819 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-whitelightstep_0003.rmap 1.5 K bytes (103 / 200 files) (363.6 K / 717.9 K bytes)
2024-12-10 20:14:01,003 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-tweakregstep_0003.rmap 4.5 K bytes (104 / 200 files) (365.1 K / 717.9 K bytes)
2024-12-10 20:14:01,282 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-spec2pipeline_0008.rmap 984 bytes (105 / 200 files) (369.5 K / 717.9 K bytes)
2024-12-10 20:14:01,559 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-sourcecatalogstep_0002.rmap 4.6 K bytes (106 / 200 files) (370.5 K / 717.9 K bytes)
2024-12-10 20:14:01,769 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-resamplestep_0002.rmap 687 bytes (107 / 200 files) (375.2 K / 717.9 K bytes)
2024-12-10 20:14:01,958 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-outlierdetectionstep_0003.rmap 940 bytes (108 / 200 files) (375.8 K / 717.9 K bytes)
2024-12-10 20:14:02,159 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-jumpstep_0005.rmap 806 bytes (109 / 200 files) (376.8 K / 717.9 K bytes)
2024-12-10 20:14:02,293 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-image2pipeline_0003.rmap 1.0 K bytes (110 / 200 files) (377.6 K / 717.9 K bytes)
2024-12-10 20:14:02,533 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-detector1pipeline_0003.rmap 1.0 K bytes (111 / 200 files) (378.6 K / 717.9 K bytes)
2024-12-10 20:14:02,781 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-darkpipeline_0002.rmap 868 bytes (112 / 200 files) (379.6 K / 717.9 K bytes)
2024-12-10 20:14:02,977 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_pars-darkcurrentstep_0001.rmap 618 bytes (113 / 200 files) (380.5 K / 717.9 K bytes)
2024-12-10 20:14:03,106 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_mask_0011.rmap 3.5 K bytes (114 / 200 files) (381.1 K / 717.9 K bytes)
2024-12-10 20:14:03,225 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_linearity_0011.rmap 2.4 K bytes (115 / 200 files) (384.6 K / 717.9 K bytes)
2024-12-10 20:14:03,339 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_ipc_0003.rmap 2.0 K bytes (116 / 200 files) (387.0 K / 717.9 K bytes)
2024-12-10 20:14:03,446 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_gain_0016.rmap 2.1 K bytes (117 / 200 files) (389.0 K / 717.9 K bytes)
2024-12-10 20:14:03,569 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_flat_0027.rmap 51.7 K bytes (118 / 200 files) (391.1 K / 717.9 K bytes)
2024-12-10 20:14:03,736 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_filteroffset_0004.rmap 1.4 K bytes (119 / 200 files) (442.8 K / 717.9 K bytes)
2024-12-10 20:14:03,885 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_extract1d_0004.rmap 842 bytes (120 / 200 files) (444.2 K / 717.9 K bytes)
2024-12-10 20:14:04,079 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_drizpars_0001.rmap 519 bytes (121 / 200 files) (445.1 K / 717.9 K bytes)
2024-12-10 20:14:04,194 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_distortion_0033.rmap 53.4 K bytes (122 / 200 files) (445.6 K / 717.9 K bytes)
2024-12-10 20:14:04,359 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_dark_0045.rmap 26.3 K bytes (123 / 200 files) (499.0 K / 717.9 K bytes)
2024-12-10 20:14:04,506 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_area_0012.rmap 33.5 K bytes (124 / 200 files) (525.3 K / 717.9 K bytes)
2024-12-10 20:14:04,655 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_apcorr_0008.rmap 4.3 K bytes (125 / 200 files) (558.8 K / 717.9 K bytes)
2024-12-10 20:14:04,783 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_abvegaoffset_0003.rmap 1.3 K bytes (126 / 200 files) (563.1 K / 717.9 K bytes)
2024-12-10 20:14:04,947 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_nircam_0297.imap 5.5 K bytes (127 / 200 files) (564.4 K / 717.9 K bytes)
2024-12-10 20:14:05,108 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_wavelengthrange_0027.rmap 929 bytes (128 / 200 files) (569.8 K / 717.9 K bytes)
2024-12-10 20:14:05,218 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_tsophot_0004.rmap 882 bytes (129 / 200 files) (570.8 K / 717.9 K bytes)
2024-12-10 20:14:05,325 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_straymask_0009.rmap 987 bytes (130 / 200 files) (571.6 K / 717.9 K bytes)
2024-12-10 20:14:05,463 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_specwcs_0042.rmap 5.8 K bytes (131 / 200 files) (572.6 K / 717.9 K bytes)
2024-12-10 20:14:05,630 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_saturation_0015.rmap 1.2 K bytes (132 / 200 files) (578.4 K / 717.9 K bytes)
2024-12-10 20:14:05,738 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_rscd_0008.rmap 1.0 K bytes (133 / 200 files) (579.6 K / 717.9 K bytes)
2024-12-10 20:14:05,891 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_resol_0006.rmap 790 bytes (134 / 200 files) (580.6 K / 717.9 K bytes)
2024-12-10 20:14:06,012 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_reset_0026.rmap 3.9 K bytes (135 / 200 files) (581.4 K / 717.9 K bytes)
2024-12-10 20:14:06,124 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_regions_0033.rmap 5.2 K bytes (136 / 200 files) (585.3 K / 717.9 K bytes)
2024-12-10 20:14:06,245 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_readnoise_0023.rmap 1.6 K bytes (137 / 200 files) (590.5 K / 717.9 K bytes)
2024-12-10 20:14:06,384 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_psfmask_0009.rmap 2.1 K bytes (138 / 200 files) (592.1 K / 717.9 K bytes)
2024-12-10 20:14:06,524 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_photom_0056.rmap 3.7 K bytes (139 / 200 files) (594.2 K / 717.9 K bytes)
2024-12-10 20:14:06,652 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pathloss_0005.rmap 866 bytes (140 / 200 files) (598.0 K / 717.9 K bytes)
2024-12-10 20:14:06,768 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-whitelightstep_0003.rmap 912 bytes (141 / 200 files) (598.9 K / 717.9 K bytes)
2024-12-10 20:14:06,882 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-tweakregstep_0003.rmap 1.8 K bytes (142 / 200 files) (599.8 K / 717.9 K bytes)
2024-12-10 20:14:07,005 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-spec3pipeline_0008.rmap 816 bytes (143 / 200 files) (601.6 K / 717.9 K bytes)
2024-12-10 20:14:07,112 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-spec2pipeline_0012.rmap 1.3 K bytes (144 / 200 files) (602.4 K / 717.9 K bytes)
2024-12-10 20:14:07,225 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-sourcecatalogstep_0003.rmap 1.9 K bytes (145 / 200 files) (603.7 K / 717.9 K bytes)
2024-12-10 20:14:07,340 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-resamplestep_0002.rmap 677 bytes (146 / 200 files) (605.6 K / 717.9 K bytes)
2024-12-10 20:14:07,454 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-resamplespecstep_0002.rmap 706 bytes (147 / 200 files) (606.3 K / 717.9 K bytes)
2024-12-10 20:14:07,562 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-outlierdetectionstep_0017.rmap 3.4 K bytes (148 / 200 files) (607.0 K / 717.9 K bytes)
2024-12-10 20:14:07,671 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-jumpstep_0009.rmap 1.4 K bytes (149 / 200 files) (610.4 K / 717.9 K bytes)
2024-12-10 20:14:07,779 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-image2pipeline_0007.rmap 983 bytes (150 / 200 files) (611.8 K / 717.9 K bytes)
2024-12-10 20:14:07,898 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-extract1dstep_0002.rmap 728 bytes (151 / 200 files) (612.8 K / 717.9 K bytes)
2024-12-10 20:14:08,008 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-emicorrstep_0002.rmap 796 bytes (152 / 200 files) (613.6 K / 717.9 K bytes)
2024-12-10 20:14:08,120 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-detector1pipeline_0009.rmap 1.4 K bytes (153 / 200 files) (614.4 K / 717.9 K bytes)
2024-12-10 20:14:08,241 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-darkpipeline_0002.rmap 860 bytes (154 / 200 files) (615.8 K / 717.9 K bytes)
2024-12-10 20:14:08,349 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_pars-darkcurrentstep_0002.rmap 683 bytes (155 / 200 files) (616.6 K / 717.9 K bytes)
2024-12-10 20:14:08,462 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_mrsxartcorr_0002.rmap 2.2 K bytes (156 / 200 files) (617.3 K / 717.9 K bytes)
2024-12-10 20:14:08,579 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_mrsptcorr_0005.rmap 2.0 K bytes (157 / 200 files) (619.5 K / 717.9 K bytes)
2024-12-10 20:14:08,685 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_mask_0023.rmap 3.5 K bytes (158 / 200 files) (621.4 K / 717.9 K bytes)
2024-12-10 20:14:08,789 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_linearity_0018.rmap 2.8 K bytes (159 / 200 files) (624.9 K / 717.9 K bytes)
2024-12-10 20:14:08,905 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_ipc_0008.rmap 700 bytes (160 / 200 files) (627.7 K / 717.9 K bytes)
2024-12-10 20:14:09,020 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_gain_0013.rmap 3.9 K bytes (161 / 200 files) (628.4 K / 717.9 K bytes)
2024-12-10 20:14:09,127 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_fringefreq_0003.rmap 1.4 K bytes (162 / 200 files) (632.4 K / 717.9 K bytes)
2024-12-10 20:14:09,242 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_fringe_0019.rmap 3.9 K bytes (163 / 200 files) (633.8 K / 717.9 K bytes)
2024-12-10 20:14:09,350 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_flat_0065.rmap 15.5 K bytes (164 / 200 files) (637.7 K / 717.9 K bytes)
2024-12-10 20:14:09,481 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_filteroffset_0025.rmap 2.5 K bytes (165 / 200 files) (653.2 K / 717.9 K bytes)
2024-12-10 20:14:09,585 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_extract1d_0020.rmap 1.4 K bytes (166 / 200 files) (655.7 K / 717.9 K bytes)
2024-12-10 20:14:09,698 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_emicorr_0003.rmap 663 bytes (167 / 200 files) (657.1 K / 717.9 K bytes)
2024-12-10 20:14:09,804 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_drizpars_0002.rmap 511 bytes (168 / 200 files) (657.7 K / 717.9 K bytes)
2024-12-10 20:14:09,917 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_distortion_0040.rmap 4.9 K bytes (169 / 200 files) (658.2 K / 717.9 K bytes)
2024-12-10 20:14:10,030 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_dark_0036.rmap 4.4 K bytes (170 / 200 files) (663.1 K / 717.9 K bytes)
2024-12-10 20:14:10,136 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_cubepar_0017.rmap 800 bytes (171 / 200 files) (667.5 K / 717.9 K bytes)
2024-12-10 20:14:10,246 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_area_0015.rmap 866 bytes (172 / 200 files) (668.3 K / 717.9 K bytes)
2024-12-10 20:14:10,352 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_apcorr_0019.rmap 5.0 K bytes (173 / 200 files) (669.2 K / 717.9 K bytes)
2024-12-10 20:14:10,474 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_abvegaoffset_0002.rmap 1.3 K bytes (174 / 200 files) (674.1 K / 717.9 K bytes)
2024-12-10 20:14:10,583 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_miri_0416.imap 5.7 K bytes (175 / 200 files) (675.4 K / 717.9 K bytes)
2024-12-10 20:14:10,699 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_trappars_0004.rmap 903 bytes (176 / 200 files) (681.1 K / 717.9 K bytes)
2024-12-10 20:14:10,808 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_trapdensity_0006.rmap 930 bytes (177 / 200 files) (682.0 K / 717.9 K bytes)
2024-12-10 20:14:10,931 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_superbias_0017.rmap 3.8 K bytes (178 / 200 files) (682.9 K / 717.9 K bytes)
2024-12-10 20:14:11,043 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_saturation_0009.rmap 779 bytes (179 / 200 files) (686.7 K / 717.9 K bytes)
2024-12-10 20:14:11,151 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_readnoise_0011.rmap 1.3 K bytes (180 / 200 files) (687.5 K / 717.9 K bytes)
2024-12-10 20:14:11,256 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_photom_0014.rmap 1.1 K bytes (181 / 200 files) (688.8 K / 717.9 K bytes)
2024-12-10 20:14:11,363 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_persat_0006.rmap 884 bytes (182 / 200 files) (689.9 K / 717.9 K bytes)
2024-12-10 20:14:11,467 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-tweakregstep_0002.rmap 850 bytes (183 / 200 files) (690.8 K / 717.9 K bytes)
2024-12-10 20:14:11,572 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-sourcecatalogstep_0001.rmap 636 bytes (184 / 200 files) (691.6 K / 717.9 K bytes)
2024-12-10 20:14:11,701 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-outlierdetectionstep_0001.rmap 654 bytes (185 / 200 files) (692.2 K / 717.9 K bytes)
2024-12-10 20:14:11,809 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-image2pipeline_0005.rmap 974 bytes (186 / 200 files) (692.9 K / 717.9 K bytes)
2024-12-10 20:14:11,916 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-detector1pipeline_0002.rmap 1.0 K bytes (187 / 200 files) (693.9 K / 717.9 K bytes)
2024-12-10 20:14:12,026 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_pars-darkpipeline_0002.rmap 856 bytes (188 / 200 files) (694.9 K / 717.9 K bytes)
2024-12-10 20:14:12,135 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_mask_0023.rmap 1.1 K bytes (189 / 200 files) (695.8 K / 717.9 K bytes)
2024-12-10 20:14:12,244 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_linearity_0015.rmap 925 bytes (190 / 200 files) (696.8 K / 717.9 K bytes)
2024-12-10 20:14:12,349 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_ipc_0003.rmap 614 bytes (191 / 200 files) (697.7 K / 717.9 K bytes)
2024-12-10 20:14:12,456 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_gain_0010.rmap 890 bytes (192 / 200 files) (698.4 K / 717.9 K bytes)
2024-12-10 20:14:12,569 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_flat_0009.rmap 1.1 K bytes (193 / 200 files) (699.2 K / 717.9 K bytes)
2024-12-10 20:14:12,681 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_distortion_0011.rmap 1.2 K bytes (194 / 200 files) (700.4 K / 717.9 K bytes)
2024-12-10 20:14:12,789 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_dark_0017.rmap 4.3 K bytes (195 / 200 files) (701.6 K / 717.9 K bytes)
2024-12-10 20:14:12,900 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_area_0010.rmap 1.2 K bytes (196 / 200 files) (705.9 K / 717.9 K bytes)
2024-12-10 20:14:13,017 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_apcorr_0004.rmap 4.0 K bytes (197 / 200 files) (707.1 K / 717.9 K bytes)
2024-12-10 20:14:13,124 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_abvegaoffset_0002.rmap 1.3 K bytes (198 / 200 files) (711.0 K / 717.9 K bytes)
2024-12-10 20:14:13,234 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_fgs_0117.imap 5.0 K bytes (199 / 200 files) (712.3 K / 717.9 K bytes)
2024-12-10 20:14:13,342 - CRDS - INFO - Fetching crds_cache/mappings/jwst/jwst_1303.pmap 580 bytes (200 / 200 files) (717.3 K / 717.9 K bytes)
2024-12-10 20:14:13,597 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:14:13,601 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf 1.1 K bytes (1 / 1 files) (0 / 1.1 K bytes)
2024-12-10 20:14:13,714 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:14:13,725 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf 1.6 K bytes (1 / 1 files) (0 / 1.6 K bytes)
2024-12-10 20:14:13,871 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:14:13,881 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:14:13,884 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf 1.1 K bytes (1 / 1 files) (0 / 1.1 K bytes)
2024-12-10 20:14:13,997 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:14:14,015 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:14:14,016 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:14:14,017 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:14:14,018 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:14:14,019 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:14:14,020 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:14:14,021 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:14:14,022 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:14:14,023 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:14:14,024 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:14:14,025 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:14:14,026 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:14:14,027 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:14:14,028 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:14:14,030 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:14:14,031 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:14:14,033 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:14:14,034 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:14:14,035 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:14:14,037 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:14:14,200 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00001_nis_uncal.fits',).
2024-12-10 20:14:14,222 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:14:14,381 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00001_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:14:14,384 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits 1.0 G bytes (1 / 7 files) (0 / 1.4 G bytes)
2024-12-10 20:14:29,570 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits 16.8 M bytes (2 / 7 files) (1.0 G / 1.4 G bytes)
2024-12-10 20:14:30,066 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits 205.5 M bytes (3 / 7 files) (1.0 G / 1.4 G bytes)
2024-12-10 20:14:33,583 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits 16.8 M bytes (4 / 7 files) (1.2 G / 1.4 G bytes)
2024-12-10 20:14:34,192 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits 16.8 M bytes (5 / 7 files) (1.3 G / 1.4 G bytes)
2024-12-10 20:14:34,760 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits 33.6 M bytes (6 / 7 files) (1.3 G / 1.4 G bytes)
2024-12-10 20:14:36,403 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits 50.4 M bytes (7 / 7 files) (1.3 G / 1.4 G bytes)
2024-12-10 20:14:41,036 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:14:41,037 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:14:41,038 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:14:41,038 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:14:41,039 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:14:41,040 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:14:41,040 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:14:41,041 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:14:41,041 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:14:41,042 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:14:41,043 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:14:41,422 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:41,432 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:14:41,433 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:14:41,434 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:14:41,574 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:41,592 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:14:41,852 - CRDS - INFO - Calibration SW Found: jwst 1.16.1 (/opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst-1.16.1.dist-info)
2024-12-10 20:14:41,974 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:14:42,138 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:42,157 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:14:42,187 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:42,188 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:42,193 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:42,281 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:14:45,790 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4239 saturated pixels
2024-12-10 20:14:45,821 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:14:45,827 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:14:45,982 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:45,983 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:14:46,119 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:46,138 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:14:46,409 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:14:46,563 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:46,651 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:14:46,652 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:14:46,653 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:14:46,653 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:14:46,653 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:14:46,654 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:14:46,654 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:14:46,655 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:14:51,093 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:14:51,244 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:51,263 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:14:51,326 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:51,327 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:51,331 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:14:51,896 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:14:52,047 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:52,048 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:14:52,197 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:52,216 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:14:52,534 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:14:52,535 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:14:52,727 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:14:52,894 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:52,984 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:14:53,734 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:14:53,885 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:14:53,980 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:14:53,981 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:14:53,992 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:14:54,016 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:14:54,221 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:15:04,536 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:15:05,924 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 34
2024-12-10 20:15:05,925 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.7034 sec
2024-12-10 20:15:06,017 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.122203
2024-12-10 20:15:06,020 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:15:06,182 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:15:06,182 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:15:06,324 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:15:06,431 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:15:06,432 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:15:06,460 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:15:06,460 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:15:07,034 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:15:12,649 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.611459493637085
2024-12-10 20:15:12,708 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:15:12,879 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:15:12,907 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:15:12,908 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:15:12,910 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:15:13,078 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00001_nis_uncal.fits>,).
2024-12-10 20:15:13,108 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:15:13,109 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:15:13,111 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:15:13,265 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00001_nis_rateints.fits
2024-12-10 20:15:13,266 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:15:13,267 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:15:13,411 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00001_nis_rate.fits
2024-12-10 20:15:13,411 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:15:13,412 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:15:13,578 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:15:13,581 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:15:13,590 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:15:13,600 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:15:13,603 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:15:13,620 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:15:13,622 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:15:13,622 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:15:13,624 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:15:13,625 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:15:13,626 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:15:13,627 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:15:13,628 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:15:13,629 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:15:13,630 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:15:13,631 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:15:13,631 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:15:13,633 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:15:13,634 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:15:13,635 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:15:13,636 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:15:13,637 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:15:13,639 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:15:13,640 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:15:13,641 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:15:13,798 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00002_nis_uncal.fits',).
2024-12-10 20:15:13,820 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:15:13,985 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00002_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:15:13,988 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:15:13,989 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:15:13,989 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:15:13,989 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:15:13,990 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:15:13,990 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:15:13,991 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:15:13,991 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:15:13,991 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:15:13,992 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:15:13,994 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:15:14,379 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:14,389 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:15:14,390 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:15:14,391 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:15:14,558 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:14,577 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:15:14,812 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:15:14,974 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:14,993 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:15:15,018 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:15,019 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:15,023 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:15,108 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:15:18,399 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4272 saturated pixels
2024-12-10 20:15:18,429 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:15:18,436 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:15:18,594 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:18,595 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:15:18,749 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:18,770 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:15:19,044 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:15:19,216 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:19,306 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:15:19,307 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:15:19,307 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:15:19,308 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:15:19,308 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:15:19,309 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:15:19,310 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:15:19,310 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:15:23,722 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:15:23,893 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:23,913 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:15:23,967 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:23,968 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:23,972 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:24,541 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:15:24,698 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:24,699 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:15:24,850 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:24,871 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:15:25,143 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:15:25,144 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:15:25,337 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:15:25,501 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:25,591 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:15:26,333 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:15:26,498 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:26,592 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:15:26,592 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:15:26,604 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:15:26,627 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:15:26,826 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:15:36,886 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:15:38,207 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 38
2024-12-10 20:15:38,208 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.3818 sec
2024-12-10 20:15:38,300 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.792016
2024-12-10 20:15:38,303 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:15:38,462 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:38,463 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:15:38,607 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:38,714 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:15:38,714 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:15:38,741 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:15:38,742 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:15:39,316 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:15:44,910 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.590129137039185
2024-12-10 20:15:44,963 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:15:45,125 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:45,154 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:15:45,154 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:15:45,156 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:15:45,301 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00002_nis_uncal.fits>,).
2024-12-10 20:15:45,332 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:15:45,333 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:15:45,335 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:15:45,486 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00002_nis_rateints.fits
2024-12-10 20:15:45,487 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:15:45,488 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:15:45,634 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00002_nis_rate.fits
2024-12-10 20:15:45,634 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:15:45,635 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:15:45,802 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:15:45,805 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:15:45,814 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:15:45,824 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:15:45,826 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:15:45,844 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:15:45,845 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:15:45,846 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:15:45,847 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:15:45,848 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:15:45,850 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:15:45,851 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:15:45,852 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:15:45,852 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:15:45,853 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:15:45,855 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:15:45,856 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:15:45,857 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:15:45,858 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:15:45,859 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:15:45,860 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:15:45,863 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:15:45,864 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:15:45,865 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:15:45,866 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:15:46,027 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00003_nis_uncal.fits',).
2024-12-10 20:15:46,048 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:15:46,214 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00003_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:15:46,217 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:15:46,218 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:15:46,218 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:15:46,219 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:15:46,220 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:15:46,221 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:15:46,221 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:15:46,221 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:15:46,222 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:15:46,223 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:15:46,224 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:15:46,592 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:46,602 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:15:46,602 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:15:46,604 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:15:46,748 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:46,768 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:15:47,000 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:15:47,164 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:47,184 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:15:47,208 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:47,208 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:47,213 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:47,295 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:15:50,397 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4611 saturated pixels
2024-12-10 20:15:50,428 - stpipe.Detector1Pipeline.saturation - INFO - Detected 2 A/D floor pixels
2024-12-10 20:15:50,433 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:15:50,606 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:50,607 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:15:50,763 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:50,784 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:15:51,054 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:15:51,220 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:51,311 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:15:51,312 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:15:51,312 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:15:51,313 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:15:51,313 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:15:51,314 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:15:51,314 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:15:51,315 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:15:55,648 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:15:55,815 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:55,837 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:15:55,889 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:55,889 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:55,893 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:15:56,452 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:15:56,614 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:56,615 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:15:56,766 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:56,787 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:15:57,055 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:15:57,055 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:15:57,245 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:15:57,417 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:57,507 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:15:58,259 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:15:58,435 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:15:58,528 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:15:58,529 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:15:58,540 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:15:58,563 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:15:58,762 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:16:08,936 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:16:10,446 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 48
2024-12-10 20:16:10,446 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.6834 sec
2024-12-10 20:16:10,534 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.089541
2024-12-10 20:16:10,537 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:16:10,711 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:16:10,712 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:16:10,874 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:16:10,984 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:16:10,984 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:16:11,012 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:16:11,013 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:16:11,581 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:16:17,193 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.60776686668396
2024-12-10 20:16:17,245 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:16:17,421 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:16:17,450 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:16:17,450 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:16:17,452 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:16:17,621 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00003_nis_uncal.fits>,).
2024-12-10 20:16:17,652 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:16:17,652 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:16:17,655 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:16:17,806 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00003_nis_rateints.fits
2024-12-10 20:16:17,807 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:16:17,809 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:16:17,955 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00003_nis_rate.fits
2024-12-10 20:16:17,956 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:16:17,956 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:16:18,125 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:16:18,127 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:16:18,137 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:16:18,146 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:16:18,149 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:16:18,167 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:16:18,168 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:16:18,169 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:16:18,170 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:16:18,171 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:16:18,172 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:16:18,173 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:16:18,174 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:16:18,175 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:16:18,176 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:16:18,177 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:16:18,178 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:16:18,179 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:16:18,181 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:16:18,182 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:16:18,183 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:16:18,184 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:16:18,186 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:16:18,187 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:16:18,188 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:16:18,348 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00004_nis_uncal.fits',).
2024-12-10 20:16:18,370 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:16:18,537 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00004_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:16:18,540 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:16:18,541 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:16:18,541 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:16:18,542 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:16:18,542 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:16:18,543 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:16:18,543 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:16:18,544 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:16:18,544 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:16:18,545 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:16:18,546 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:16:18,918 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:18,929 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:16:18,929 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:16:18,931 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:16:19,080 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:19,099 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:16:19,321 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:16:19,491 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:19,511 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:16:19,535 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:19,536 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:19,540 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:19,625 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:16:22,685 - stpipe.Detector1Pipeline.saturation - INFO - Detected 5030 saturated pixels
2024-12-10 20:16:22,715 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:16:22,721 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:16:22,894 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:22,895 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:16:23,065 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:23,085 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:16:23,353 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:16:23,529 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:23,620 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:16:23,621 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:16:23,621 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:16:23,622 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:16:23,622 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:16:23,622 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:16:23,623 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:16:23,623 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:16:27,922 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:16:28,091 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:28,111 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:16:28,156 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:28,157 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:28,161 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:28,724 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:16:28,900 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:28,901 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:16:29,072 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:29,093 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:16:29,359 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:16:29,359 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:16:29,541 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:16:29,716 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:29,805 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:16:30,617 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:16:30,791 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:30,884 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:16:30,885 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:16:30,895 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:16:30,918 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:16:31,119 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:16:41,233 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:16:42,694 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 46
2024-12-10 20:16:42,694 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.5749 sec
2024-12-10 20:16:42,781 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.978951
2024-12-10 20:16:42,784 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:16:42,957 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:42,958 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:16:43,129 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:43,238 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:16:43,238 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:16:43,267 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:16:43,267 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:16:43,836 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:16:49,320 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.48036003112793
2024-12-10 20:16:49,379 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:16:49,539 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:49,568 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:16:49,569 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:16:49,570 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:16:49,728 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00004_nis_uncal.fits>,).
2024-12-10 20:16:49,759 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:16:49,760 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:16:49,762 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:16:49,915 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00004_nis_rateints.fits
2024-12-10 20:16:49,916 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:16:49,918 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:16:50,061 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00004_nis_rate.fits
2024-12-10 20:16:50,061 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:16:50,062 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:16:50,220 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:16:50,222 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:16:50,232 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:16:50,241 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:16:50,244 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:16:50,261 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:16:50,262 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:16:50,263 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:16:50,264 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:16:50,265 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:16:50,266 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:16:50,266 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:16:50,267 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:16:50,268 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:16:50,269 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:16:50,270 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:16:50,271 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:16:50,272 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:16:50,273 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:16:50,274 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:16:50,275 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:16:50,277 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:16:50,278 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:16:50,279 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:16:50,280 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:16:50,436 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00005_nis_uncal.fits',).
2024-12-10 20:16:50,459 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:16:50,616 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00005_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:16:50,619 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:16:50,620 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:16:50,621 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:16:50,621 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:16:50,622 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:16:50,622 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:16:50,623 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:16:50,624 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:16:50,624 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:16:50,625 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:16:50,626 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:16:50,987 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:50,997 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:16:50,998 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:16:50,999 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:16:51,151 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:51,171 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:16:51,398 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:16:51,559 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:51,579 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:16:51,604 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:51,604 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:51,608 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:16:51,691 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:16:54,715 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4901 saturated pixels
2024-12-10 20:16:54,745 - stpipe.Detector1Pipeline.saturation - INFO - Detected 3 A/D floor pixels
2024-12-10 20:16:54,751 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:16:54,913 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:54,913 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:16:55,062 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:55,081 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:16:55,345 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:16:55,500 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:16:55,590 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:16:55,591 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:16:55,591 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:16:55,591 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:16:55,592 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:16:55,592 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:16:55,594 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:16:55,594 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:16:59,855 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:17:00,025 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:00,044 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:17:00,090 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:00,091 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:00,095 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:00,664 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:17:00,825 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:00,826 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:17:00,989 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:01,008 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:17:01,292 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:17:01,293 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:17:01,469 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:17:01,648 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:01,741 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:17:02,526 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:17:02,685 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:02,776 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:17:02,777 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:17:02,788 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:17:02,810 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:17:03,008 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:17:13,150 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:17:14,536 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 41
2024-12-10 20:17:14,537 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.5288 sec
2024-12-10 20:17:14,623 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.928194
2024-12-10 20:17:14,626 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:17:14,790 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:14,790 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:17:14,956 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:15,068 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:17:15,068 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:17:15,097 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:17:15,097 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:17:15,667 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:17:21,362 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.691061496734619
2024-12-10 20:17:21,414 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:17:21,572 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:21,601 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:17:21,601 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:17:21,603 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:17:21,762 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00005_nis_uncal.fits>,).
2024-12-10 20:17:21,793 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:17:21,794 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:17:21,795 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:17:21,944 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00005_nis_rateints.fits
2024-12-10 20:17:21,945 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:17:21,947 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:17:22,093 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00005_nis_rate.fits
2024-12-10 20:17:22,094 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:17:22,094 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:17:22,252 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:17:22,255 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:17:22,264 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:17:22,273 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:17:22,276 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:17:22,293 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:17:22,294 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:17:22,295 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:17:22,296 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:17:22,297 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:17:22,298 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:17:22,299 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:17:22,300 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:17:22,301 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:17:22,302 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:17:22,303 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:17:22,304 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:17:22,305 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:17:22,306 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:17:22,307 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:17:22,307 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:17:22,309 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:17:22,310 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:17:22,312 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:17:22,312 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:17:22,472 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00006_nis_uncal.fits',).
2024-12-10 20:17:22,493 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:17:22,649 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00006_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:17:22,652 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:17:22,653 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:17:22,654 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:17:22,654 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:17:22,655 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:17:22,655 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:17:22,656 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:17:22,656 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:17:22,657 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:17:22,658 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:17:22,659 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:17:23,027 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:23,037 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:17:23,037 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:17:23,039 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:17:23,202 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:23,222 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:17:23,449 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:17:23,625 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:23,644 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:17:23,668 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:23,669 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:23,673 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:23,755 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:17:26,779 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4479 saturated pixels
2024-12-10 20:17:26,809 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:17:26,815 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:17:26,977 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:26,978 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:17:27,129 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:27,149 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:17:27,421 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:17:27,583 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:27,674 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:17:27,674 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:17:27,675 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:17:27,675 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:17:27,676 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:17:27,676 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:17:27,676 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:17:27,677 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:17:31,950 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:17:32,110 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:32,130 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:17:32,176 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:32,177 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:32,180 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:32,739 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:17:32,896 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:32,896 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:17:33,041 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:33,061 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:17:33,328 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:17:33,329 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:17:33,509 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:17:33,666 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:33,755 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:17:34,524 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:17:34,680 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:34,771 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:17:34,771 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:17:34,783 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:17:34,806 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:17:35,001 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:17:45,110 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:17:46,364 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 41
2024-12-10 20:17:46,365 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.3625 sec
2024-12-10 20:17:46,451 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.760849
2024-12-10 20:17:46,454 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:17:46,617 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:46,618 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:17:46,777 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:46,887 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:17:46,887 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:17:46,915 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:17:46,915 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:17:47,489 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:17:53,126 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.63278603553772
2024-12-10 20:17:53,177 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:17:53,338 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:53,366 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:17:53,366 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:17:53,368 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:17:53,524 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00006_nis_uncal.fits>,).
2024-12-10 20:17:53,555 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:17:53,555 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:17:53,557 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:17:53,706 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00006_nis_rateints.fits
2024-12-10 20:17:53,707 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:17:53,709 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:17:53,851 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00006_nis_rate.fits
2024-12-10 20:17:53,852 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:17:53,853 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:17:54,019 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:17:54,022 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:17:54,031 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:17:54,041 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:17:54,043 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:17:54,061 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:17:54,062 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:17:54,063 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:17:54,064 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:17:54,065 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:17:54,065 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:17:54,066 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:17:54,067 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:17:54,068 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:17:54,069 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:17:54,070 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:17:54,071 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:17:54,072 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:17:54,073 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:17:54,074 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:17:54,075 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:17:54,076 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:17:54,078 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:17:54,079 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:17:54,079 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:17:54,240 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00007_nis_uncal.fits',).
2024-12-10 20:17:54,264 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:17:54,431 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00007_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:17:54,434 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:17:54,435 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:17:54,436 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:17:54,436 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:17:54,437 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:17:54,437 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:17:54,437 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:17:54,438 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:17:54,438 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:17:54,439 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:17:54,441 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:17:54,814 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:54,824 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:17:54,824 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:17:54,826 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:17:54,971 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:54,991 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:17:55,214 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:17:55,382 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:55,403 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:17:55,427 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:55,427 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:55,431 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:17:55,517 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:17:58,559 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4366 saturated pixels
2024-12-10 20:17:58,589 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:17:58,595 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:17:58,760 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:58,761 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:17:58,908 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:58,928 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:17:59,196 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:17:59,374 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:17:59,465 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:17:59,465 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:17:59,466 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:17:59,466 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:17:59,466 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:17:59,467 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:17:59,467 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:17:59,468 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:18:03,777 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:18:03,938 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:03,958 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:18:04,004 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:04,005 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:04,009 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:04,579 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:18:04,741 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:04,742 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:18:04,889 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:04,908 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:18:05,175 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:18:05,176 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:18:05,359 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:18:05,525 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:05,615 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:18:06,339 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:18:06,522 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:06,618 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:18:06,619 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:18:06,630 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:18:06,654 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:18:06,857 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:18:16,918 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:18:18,342 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 45
2024-12-10 20:18:18,343 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.4855 sec
2024-12-10 20:18:18,429 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.895696
2024-12-10 20:18:18,432 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:18:18,591 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:18,592 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:18:18,737 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:18,845 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:18:18,846 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:18:18,874 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:18:18,874 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:18:19,436 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:18:25,055 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.614948987960815
2024-12-10 20:18:25,106 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:18:25,272 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:25,300 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:18:25,301 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:18:25,303 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:18:25,451 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00007_nis_uncal.fits>,).
2024-12-10 20:18:25,482 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:18:25,482 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:18:25,484 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:18:25,633 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00007_nis_rateints.fits
2024-12-10 20:18:25,634 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:18:25,636 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:18:25,779 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00007_nis_rate.fits
2024-12-10 20:18:25,780 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:18:25,780 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:18:25,948 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:18:25,950 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:18:25,960 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:18:25,969 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:18:25,972 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:18:25,990 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:18:25,991 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:18:25,992 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:18:25,993 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:18:25,994 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:18:25,995 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:18:25,996 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:18:25,997 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:18:25,998 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:18:25,999 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:18:26,000 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:18:26,002 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:18:26,003 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:18:26,003 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:18:26,005 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:18:26,006 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:18:26,008 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:18:26,009 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:18:26,010 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:18:26,011 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:18:26,170 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00008_nis_uncal.fits',).
2024-12-10 20:18:26,193 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:18:26,358 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00008_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:18:26,361 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:18:26,362 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:18:26,363 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:18:26,363 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:18:26,364 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:18:26,365 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:18:26,365 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:18:26,366 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:18:26,366 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:18:26,367 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:18:26,368 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:18:26,750 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:26,760 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:18:26,760 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:18:26,762 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:18:26,916 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:26,936 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:18:27,157 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:18:27,323 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:27,343 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:18:27,367 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:27,368 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:27,372 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:27,458 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:18:30,488 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4369 saturated pixels
2024-12-10 20:18:30,519 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:18:30,525 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:18:30,699 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:30,699 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:18:30,857 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:30,877 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:18:31,146 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:18:31,314 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:31,406 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:18:31,407 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:18:31,407 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:18:31,408 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:18:31,408 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:18:31,408 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:18:31,409 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:18:31,409 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:18:35,703 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:18:35,866 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:35,886 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:18:35,938 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:35,939 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:35,943 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:36,519 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:18:36,689 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:36,689 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:18:36,853 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:36,872 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:18:37,140 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:18:37,140 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:18:37,323 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:18:37,504 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:37,595 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:18:38,346 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:18:38,513 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:38,604 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:18:38,605 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:18:38,616 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:18:38,639 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:18:38,836 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:18:49,268 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:18:50,696 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 53
2024-12-10 20:18:50,697 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.8595 sec
2024-12-10 20:18:50,787 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.264379
2024-12-10 20:18:50,790 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:18:50,950 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:50,951 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:18:51,105 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:51,212 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:18:51,213 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:18:51,240 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:18:51,240 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:18:51,803 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:18:57,391 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.584282159805298
2024-12-10 20:18:57,443 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:18:57,601 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:57,629 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:18:57,630 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:18:57,631 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:18:57,789 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00008_nis_uncal.fits>,).
2024-12-10 20:18:57,820 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:18:57,821 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:18:57,822 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:18:57,973 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00008_nis_rateints.fits
2024-12-10 20:18:57,974 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:18:57,976 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:18:58,119 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00008_nis_rate.fits
2024-12-10 20:18:58,120 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:18:58,121 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:18:58,288 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:18:58,291 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:18:58,300 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:18:58,309 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:18:58,312 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:18:58,330 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:18:58,331 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:18:58,332 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:18:58,334 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:18:58,334 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:18:58,335 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:18:58,336 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:18:58,337 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:18:58,339 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:18:58,340 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:18:58,341 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:18:58,342 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:18:58,343 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:18:58,344 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:18:58,345 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:18:58,346 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:18:58,348 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:18:58,350 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:18:58,353 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:18:58,354 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:18:58,521 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00009_nis_uncal.fits',).
2024-12-10 20:18:58,542 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:18:58,707 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00009_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:18:58,710 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:18:58,711 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:18:58,712 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:18:58,712 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:18:58,713 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:18:58,713 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:18:58,714 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:18:58,714 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:18:58,715 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:18:58,715 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:18:58,717 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:18:59,093 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:18:59,103 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:18:59,103 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:18:59,105 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:18:59,255 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:18:59,275 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:18:59,500 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:18:59,662 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:18:59,682 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:18:59,706 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:59,707 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:59,711 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:18:59,796 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:19:02,839 - stpipe.Detector1Pipeline.saturation - INFO - Detected 5198 saturated pixels
2024-12-10 20:19:02,870 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:19:02,875 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:19:03,041 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:03,042 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:19:03,188 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:03,208 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:19:03,477 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:19:03,638 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:03,730 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:19:03,731 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:19:03,731 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:19:03,732 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:19:03,732 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:19:03,733 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:19:03,733 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:19:03,733 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:19:07,971 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:19:08,137 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:08,156 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:19:08,209 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:08,210 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:08,214 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:08,771 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:19:08,935 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:08,935 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:19:09,086 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:09,107 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:19:09,375 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:19:09,375 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:19:09,559 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:19:09,718 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:09,809 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:19:10,636 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:19:10,796 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:10,891 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:19:10,892 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:19:10,903 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:19:10,926 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:19:11,125 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:19:21,480 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:19:22,825 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 39
2024-12-10 20:19:22,826 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.7 sec
2024-12-10 20:19:22,913 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.105778
2024-12-10 20:19:22,916 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:19:23,089 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:23,089 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:19:23,242 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:23,352 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:19:23,352 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:19:23,380 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:19:23,381 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:19:23,947 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:19:29,517 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.56575083732605
2024-12-10 20:19:29,570 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:19:29,731 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:29,760 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:19:29,760 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:19:29,763 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:19:29,917 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00009_nis_uncal.fits>,).
2024-12-10 20:19:29,948 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:19:29,948 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:19:29,950 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:19:30,103 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00009_nis_rateints.fits
2024-12-10 20:19:30,103 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:19:30,105 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:19:30,249 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00009_nis_rate.fits
2024-12-10 20:19:30,249 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:19:30,250 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:19:30,410 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:19:30,413 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:19:30,423 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:19:30,432 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:19:30,435 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:19:30,453 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:19:30,453 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:19:30,455 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:19:30,456 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:19:30,457 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:19:30,458 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:19:30,459 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:19:30,461 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:19:30,461 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:19:30,463 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:19:30,464 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:19:30,465 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:19:30,467 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:19:30,468 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:19:30,469 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:19:30,469 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:19:30,471 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:19:30,473 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:19:30,475 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:19:30,475 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:19:30,634 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00010_nis_uncal.fits',).
2024-12-10 20:19:30,657 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:19:30,814 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00010_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:19:30,818 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:19:30,818 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:19:30,819 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:19:30,819 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:19:30,820 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:19:30,820 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:19:30,821 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:19:30,821 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:19:30,821 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:19:30,822 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:19:30,823 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:19:31,185 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:31,195 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:19:31,196 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:19:31,197 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:19:31,355 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:31,375 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:19:31,600 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:19:31,767 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:31,788 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:19:31,812 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:31,813 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:31,817 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:31,899 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:19:34,919 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4890 saturated pixels
2024-12-10 20:19:34,949 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:19:34,954 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:19:35,129 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:35,130 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:19:35,286 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:35,306 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:19:35,578 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:19:35,745 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:35,834 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:19:35,834 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:19:35,835 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:19:35,835 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:19:35,836 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:19:35,836 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:19:35,837 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:19:35,838 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:19:40,089 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:19:40,254 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:40,274 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:19:40,326 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:40,327 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:40,330 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:19:40,879 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:19:41,039 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:41,039 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:19:41,191 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:41,214 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:19:41,477 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:19:41,478 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:19:41,657 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:19:41,816 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:41,905 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:19:42,697 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:19:42,857 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:42,948 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:19:42,949 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:19:42,960 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:19:42,983 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:19:43,177 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:19:53,349 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:19:54,600 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 42
2024-12-10 20:19:54,601 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.4226 sec
2024-12-10 20:19:54,686 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.819323
2024-12-10 20:19:54,689 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:19:54,852 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:54,853 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:19:54,998 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:19:55,107 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:19:55,107 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:19:55,135 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:19:55,135 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:19:55,696 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:20:01,334 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.634335994720459
2024-12-10 20:20:01,387 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:20:01,551 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:20:01,579 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:20:01,580 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:20:01,582 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:20:01,747 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00010_nis_uncal.fits>,).
2024-12-10 20:20:01,778 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:20:01,778 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:20:01,780 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:20:01,933 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00010_nis_rateints.fits
2024-12-10 20:20:01,933 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:20:01,936 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:20:02,081 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00010_nis_rate.fits
2024-12-10 20:20:02,082 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:20:02,082 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:20:02,252 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:20:02,255 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:20:02,265 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:20:02,274 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:20:02,276 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:20:02,294 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:20:02,295 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:20:02,296 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:20:02,297 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:20:02,298 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:20:02,299 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:20:02,300 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:20:02,301 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:20:02,302 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:20:02,303 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:20:02,303 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:20:02,304 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:20:02,305 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:20:02,306 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:20:02,307 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:20:02,308 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:20:02,310 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:20:02,313 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:20:02,314 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:20:02,315 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:20:02,482 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00011_nis_uncal.fits',).
2024-12-10 20:20:02,503 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:20:02,669 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00011_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:20:02,672 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:20:02,673 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:20:02,673 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:20:02,673 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:20:02,674 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:20:02,674 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:20:02,675 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:20:02,676 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:20:02,676 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:20:02,677 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:20:02,678 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:20:03,059 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:03,069 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:20:03,069 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:20:03,071 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:20:03,228 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:03,248 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:20:03,481 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:20:03,648 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:03,668 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:20:03,692 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:03,692 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:03,696 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:03,782 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:20:06,945 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4408 saturated pixels
2024-12-10 20:20:06,976 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:20:06,981 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:20:07,148 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:07,149 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:20:07,299 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:07,319 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:20:07,590 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:20:07,754 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:07,856 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:20:07,857 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:20:07,857 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:20:07,858 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:20:07,858 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:20:07,859 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:20:07,859 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:20:07,859 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:20:12,122 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:20:12,294 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:12,314 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:20:12,368 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:12,369 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:12,373 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:12,942 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:20:13,123 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:13,124 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:20:13,301 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:13,320 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:20:13,591 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:20:13,592 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:20:13,776 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:20:13,942 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:14,035 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:20:14,768 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:20:14,951 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:15,045 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:20:15,045 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:20:15,056 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:20:15,080 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:20:15,278 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:20:25,647 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:20:26,911 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 41
2024-12-10 20:20:26,911 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.6332 sec
2024-12-10 20:20:26,999 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.035657
2024-12-10 20:20:27,002 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:20:27,168 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:27,170 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:20:27,316 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:27,425 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:20:27,425 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:20:27,453 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:20:27,453 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:20:28,012 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:20:33,487 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.471820592880249
2024-12-10 20:20:33,540 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:20:33,709 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:33,737 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:20:33,738 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:20:33,739 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:20:33,895 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00011_nis_uncal.fits>,).
2024-12-10 20:20:33,925 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:20:33,926 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:20:33,927 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:20:34,078 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00011_nis_rateints.fits
2024-12-10 20:20:34,079 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:20:34,081 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:20:34,225 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00011_nis_rate.fits
2024-12-10 20:20:34,226 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:20:34,226 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:20:34,392 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:20:34,395 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:20:34,404 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:20:34,413 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:20:34,416 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:20:34,434 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:20:34,435 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:20:34,436 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:20:34,437 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:20:34,438 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:20:34,438 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:20:34,439 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:20:34,440 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:20:34,441 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:20:34,442 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:20:34,443 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:20:34,444 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:20:34,445 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:20:34,445 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:20:34,446 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:20:34,447 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:20:34,449 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:20:34,450 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:20:34,451 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:20:34,452 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:20:34,614 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00012_nis_uncal.fits',).
2024-12-10 20:20:34,636 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:20:34,802 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00012_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:20:34,805 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:20:34,806 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:20:34,806 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:20:34,807 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:20:34,807 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:20:34,808 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:20:34,808 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:20:34,809 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:20:34,809 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:20:34,809 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:20:34,811 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:20:35,186 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:35,196 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:20:35,196 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:20:35,198 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:20:35,351 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:35,371 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:20:35,594 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:20:35,759 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:35,779 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:20:35,803 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:35,804 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:35,808 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:35,893 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:20:39,079 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4034 saturated pixels
2024-12-10 20:20:39,109 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:20:39,115 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:20:39,274 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:39,275 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:20:39,427 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:39,447 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:20:39,715 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:20:39,878 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:39,969 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:20:39,970 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:20:39,970 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:20:39,971 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:20:39,971 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:20:39,972 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:20:39,972 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:20:39,972 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:20:44,298 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:20:44,461 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:44,481 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:20:44,533 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:44,534 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:44,538 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:20:45,097 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:20:45,258 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:45,259 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:20:45,408 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:45,429 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:20:45,697 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:20:45,698 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:20:45,879 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:20:46,041 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:46,131 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:20:46,845 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:20:47,023 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:47,117 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:20:47,118 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:20:47,129 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:20:47,153 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:20:47,351 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:20:57,693 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:20:58,880 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 37
2024-12-10 20:20:58,881 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.5291 sec
2024-12-10 20:20:58,967 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.934483
2024-12-10 20:20:58,970 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:20:59,134 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:59,135 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:20:59,288 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:20:59,397 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:20:59,398 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:20:59,425 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:20:59,425 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:20:59,982 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:21:05,476 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.489960670471191
2024-12-10 20:21:05,528 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:21:05,696 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:21:05,725 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:21:05,726 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:21:05,728 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:21:05,898 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00012_nis_uncal.fits>,).
2024-12-10 20:21:05,928 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:21:05,929 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:21:05,930 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:21:06,082 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00012_nis_rateints.fits
2024-12-10 20:21:06,082 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:21:06,084 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:21:06,229 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00012_nis_rate.fits
2024-12-10 20:21:06,229 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:21:06,230 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:21:06,398 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:21:06,401 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:21:06,411 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:21:06,420 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:21:06,422 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:21:06,440 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:21:06,441 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:21:06,442 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:21:06,443 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:21:06,444 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:21:06,445 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:21:06,446 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:21:06,447 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:21:06,448 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:21:06,449 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:21:06,450 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:21:06,452 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:21:06,453 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:21:06,454 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:21:06,455 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:21:06,456 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:21:06,458 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:21:06,459 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:21:06,461 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:21:06,462 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:21:06,634 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00013_nis_uncal.fits',).
2024-12-10 20:21:06,656 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:21:06,822 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00013_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:21:06,825 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:21:06,826 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:21:06,826 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:21:06,827 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:21:06,827 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:21:06,828 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:21:06,828 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:21:06,828 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:21:06,829 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:21:06,830 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:21:06,832 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:21:07,215 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:07,225 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:21:07,226 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:21:07,227 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:21:07,389 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:07,410 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:21:07,636 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:21:07,814 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:07,834 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:21:07,859 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:07,860 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:07,864 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:07,950 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:21:11,119 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4411 saturated pixels
2024-12-10 20:21:11,150 - stpipe.Detector1Pipeline.saturation - INFO - Detected 2 A/D floor pixels
2024-12-10 20:21:11,156 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:21:11,319 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:11,320 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:21:11,473 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:11,494 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:21:11,765 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:21:11,938 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:12,033 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:21:12,034 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:21:12,034 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:21:12,034 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:21:12,035 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:21:12,035 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:21:12,036 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:21:12,036 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:21:16,405 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:21:16,573 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:16,593 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:21:16,646 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:16,647 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:16,651 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:17,220 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:21:17,394 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:17,395 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:21:17,561 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:17,580 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:21:17,849 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:21:17,849 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:21:18,032 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:21:18,206 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:18,296 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:21:19,083 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:21:19,260 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:19,354 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:21:19,355 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:21:19,366 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:21:19,389 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:21:19,588 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:21:29,845 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:21:31,155 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 39
2024-12-10 20:21:31,156 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.5673 sec
2024-12-10 20:21:31,245 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 11.974618
2024-12-10 20:21:31,248 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:21:31,441 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:31,442 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:21:31,632 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:31,746 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:21:31,747 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:21:31,775 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:21:31,776 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:21:32,349 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:21:37,831 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.478518962860107
2024-12-10 20:21:37,884 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:21:38,065 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:38,093 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:21:38,094 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:21:38,096 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:21:38,277 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00013_nis_uncal.fits>,).
2024-12-10 20:21:38,308 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:21:38,309 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:21:38,311 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:21:38,471 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00013_nis_rateints.fits
2024-12-10 20:21:38,472 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:21:38,474 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:21:38,622 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00013_nis_rate.fits
2024-12-10 20:21:38,622 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:21:38,623 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:21:38,792 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:21:38,794 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:21:38,804 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:21:38,813 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:21:38,816 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:21:38,834 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:21:38,835 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:21:38,836 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:21:38,837 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:21:38,838 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:21:38,839 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:21:38,840 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:21:38,841 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:21:38,842 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:21:38,843 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:21:38,844 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:21:38,846 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:21:38,847 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:21:38,848 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:21:38,849 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:21:38,850 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:21:38,852 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:21:38,853 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:21:38,855 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:21:38,856 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:21:39,038 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00014_nis_uncal.fits',).
2024-12-10 20:21:39,061 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:21:39,229 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00014_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:21:39,232 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:21:39,233 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:21:39,233 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:21:39,234 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:21:39,235 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:21:39,235 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:21:39,236 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:21:39,237 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:21:39,238 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:21:39,238 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:21:39,240 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:21:39,641 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:39,651 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:21:39,651 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:21:39,653 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:21:39,833 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:39,852 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:21:40,082 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:21:40,263 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:40,282 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:21:40,307 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:40,308 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:40,311 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:40,398 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:21:43,605 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4603 saturated pixels
2024-12-10 20:21:43,636 - stpipe.Detector1Pipeline.saturation - INFO - Detected 2 A/D floor pixels
2024-12-10 20:21:43,641 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:21:43,813 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:43,814 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:21:43,983 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:44,003 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:21:44,271 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:21:44,450 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:44,542 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:21:44,542 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:21:44,543 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:21:44,543 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:21:44,544 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:21:44,544 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:21:44,544 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:21:44,545 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:21:48,856 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:21:49,029 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:49,050 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:21:49,103 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:49,104 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:49,107 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:21:49,668 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:21:49,831 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:49,832 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:21:49,985 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:50,005 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:21:50,273 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:21:50,274 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:21:50,458 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:21:50,635 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:50,726 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:21:51,496 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:21:51,670 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:21:51,763 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:21:51,764 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:21:51,775 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:21:51,798 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:21:51,995 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:22:02,404 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:22:03,633 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 37
2024-12-10 20:22:03,634 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.6382 sec
2024-12-10 20:22:03,722 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.041989
2024-12-10 20:22:03,725 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:22:03,907 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:22:03,907 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:22:04,087 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:22:04,198 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:22:04,198 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:22:04,226 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:22:04,227 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:22:04,798 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:22:10,297 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.4944987297058105
2024-12-10 20:22:10,350 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:22:10,529 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:22:10,558 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:22:10,559 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:22:10,561 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:22:10,739 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00014_nis_uncal.fits>,).
2024-12-10 20:22:10,770 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:22:10,771 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:22:10,773 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:22:10,925 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00014_nis_rateints.fits
2024-12-10 20:22:10,925 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:22:10,928 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:22:11,074 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00014_nis_rate.fits
2024-12-10 20:22:11,074 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:22:11,075 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:22:11,242 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:22:11,245 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:22:11,255 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:22:11,264 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:22:11,267 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:22:11,286 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:22:11,287 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:22:11,288 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:22:11,289 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:22:11,290 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:22:11,291 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:22:11,292 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:22:11,294 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:22:11,295 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:22:11,296 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:22:11,297 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:22:11,298 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:22:11,300 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:22:11,301 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:22:11,302 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:22:11,303 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:22:11,305 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:22:11,306 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:22:11,307 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:22:11,308 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:22:11,488 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00015_nis_uncal.fits',).
2024-12-10 20:22:11,511 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:22:11,677 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00015_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:22:11,680 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:22:11,681 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:22:11,681 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:22:11,682 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:22:11,682 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:22:11,683 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:22:11,683 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:22:11,684 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:22:11,684 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:22:11,685 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:22:11,687 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:22:12,085 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:12,094 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:22:12,095 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:22:12,096 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:22:12,277 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:12,297 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:22:12,523 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:22:12,704 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:12,723 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:22:12,748 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:12,749 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:12,753 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:12,837 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:22:15,890 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4798 saturated pixels
2024-12-10 20:22:15,920 - stpipe.Detector1Pipeline.saturation - INFO - Detected 1 A/D floor pixels
2024-12-10 20:22:15,925 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:22:16,105 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:16,106 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:22:16,279 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:16,299 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:22:16,569 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:22:16,750 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:16,840 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:22:16,841 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:22:16,841 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:22:16,842 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:22:16,842 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:22:16,843 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:22:16,843 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:22:16,844 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:22:21,194 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:22:21,378 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:21,397 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:22:21,451 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:21,451 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:21,455 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:22,024 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:22:22,206 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:22,207 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:22:22,386 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:22,406 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:22:22,674 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:22:22,675 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:22:22,859 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:22:23,040 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:23,131 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:22:23,924 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:22:24,106 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:24,198 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:22:24,199 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:22:24,210 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:22:24,233 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:22:24,433 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:22:34,851 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:22:36,240 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 43
2024-12-10 20:22:36,240 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.8065 sec
2024-12-10 20:22:36,327 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.211697
2024-12-10 20:22:36,330 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:22:36,511 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:36,512 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:22:36,693 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:36,803 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:22:36,804 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:22:36,831 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:22:36,832 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:22:37,403 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:22:42,955 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.54801869392395
2024-12-10 20:22:43,008 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:22:43,185 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:43,215 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:22:43,216 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:22:43,217 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:22:43,391 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00015_nis_uncal.fits>,).
2024-12-10 20:22:43,422 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:22:43,423 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:22:43,424 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:22:43,576 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00015_nis_rateints.fits
2024-12-10 20:22:43,577 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:22:43,579 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:22:43,724 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00015_nis_rate.fits
2024-12-10 20:22:43,725 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:22:43,725 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:22:43,889 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:22:43,892 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:22:43,901 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:22:43,911 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:22:43,913 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:22:43,931 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:22:43,932 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:22:43,933 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:22:43,934 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:22:43,936 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:22:43,937 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:22:43,938 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:22:43,939 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:22:43,941 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:22:43,942 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:22:43,943 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:22:43,944 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:22:43,945 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:22:43,946 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:22:43,947 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:22:43,948 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:22:43,950 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:22:43,951 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:22:43,953 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:22:43,954 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:22:44,130 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00016_nis_uncal.fits',).
2024-12-10 20:22:44,151 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:22:44,309 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00016_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:22:44,312 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:22:44,313 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:22:44,314 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:22:44,314 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:22:44,315 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:22:44,315 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:22:44,315 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:22:44,316 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:22:44,316 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:22:44,317 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:22:44,319 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:22:44,707 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:44,717 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:22:44,718 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:22:44,720 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:22:44,900 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:44,920 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:22:45,149 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:22:45,330 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:45,349 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:22:45,373 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:45,374 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:45,378 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:45,463 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:22:48,667 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4919 saturated pixels
2024-12-10 20:22:48,698 - stpipe.Detector1Pipeline.saturation - INFO - Detected 2 A/D floor pixels
2024-12-10 20:22:48,703 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:22:48,884 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:48,885 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:22:49,066 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:49,086 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:22:49,359 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:22:49,540 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:49,632 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:22:49,632 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:22:49,633 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:22:49,633 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:22:49,634 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:22:49,634 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:22:49,635 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:22:49,635 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:22:53,973 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:22:54,157 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:54,177 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:22:54,223 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:54,224 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:54,228 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:22:54,792 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:22:54,977 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:54,977 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:22:55,160 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:55,180 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:22:55,448 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:22:55,449 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:22:55,622 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:22:55,805 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:55,896 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:22:56,685 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:22:56,866 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:22:56,961 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:22:56,962 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:22:56,973 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:22:56,996 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:22:57,193 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:23:07,536 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:23:09,033 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 55
2024-12-10 20:23:09,033 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.8392 sec
2024-12-10 20:23:09,121 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.244762
2024-12-10 20:23:09,124 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:23:09,305 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:23:09,305 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:23:09,485 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:23:09,594 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:23:09,595 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:23:09,623 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:23:09,624 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:23:10,193 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:23:15,669 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.47261381149292
2024-12-10 20:23:15,724 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:23:15,906 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:23:15,935 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:23:15,936 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:23:15,938 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:23:16,119 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00016_nis_uncal.fits>,).
2024-12-10 20:23:16,151 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:23:16,152 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:23:16,153 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:23:16,306 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00016_nis_rateints.fits
2024-12-10 20:23:16,306 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:23:16,308 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:23:16,454 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00016_nis_rate.fits
2024-12-10 20:23:16,455 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:23:16,455 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:23:16,626 - CRDS - ERROR - Error determining best reference for 'pars-darkcurrentstep' = No match found.
2024-12-10 20:23:16,629 - stpipe - INFO - PARS-CHARGEMIGRATIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-chargemigrationstep_0018.asdf
2024-12-10 20:23:16,638 - stpipe - INFO - PARS-JUMPSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-jumpstep_0087.asdf
2024-12-10 20:23:16,648 - CRDS - ERROR - Error determining best reference for 'pars-cleanflickernoisestep' = Unknown reference type 'pars-cleanflickernoisestep'
2024-12-10 20:23:16,650 - stpipe - INFO - PARS-DETECTOR1PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-detector1pipeline_0001.asdf
2024-12-10 20:23:16,668 - stpipe.Detector1Pipeline - INFO - Detector1Pipeline instance created.
2024-12-10 20:23:16,669 - stpipe.Detector1Pipeline.group_scale - INFO - GroupScaleStep instance created.
2024-12-10 20:23:16,670 - stpipe.Detector1Pipeline.dq_init - INFO - DQInitStep instance created.
2024-12-10 20:23:16,672 - stpipe.Detector1Pipeline.emicorr - INFO - EmiCorrStep instance created.
2024-12-10 20:23:16,673 - stpipe.Detector1Pipeline.saturation - INFO - SaturationStep instance created.
2024-12-10 20:23:16,674 - stpipe.Detector1Pipeline.ipc - INFO - IPCStep instance created.
2024-12-10 20:23:16,675 - stpipe.Detector1Pipeline.superbias - INFO - SuperBiasStep instance created.
2024-12-10 20:23:16,676 - stpipe.Detector1Pipeline.refpix - INFO - RefPixStep instance created.
2024-12-10 20:23:16,677 - stpipe.Detector1Pipeline.rscd - INFO - RscdStep instance created.
2024-12-10 20:23:16,679 - stpipe.Detector1Pipeline.firstframe - INFO - FirstFrameStep instance created.
2024-12-10 20:23:16,679 - stpipe.Detector1Pipeline.lastframe - INFO - LastFrameStep instance created.
2024-12-10 20:23:16,681 - stpipe.Detector1Pipeline.linearity - INFO - LinearityStep instance created.
2024-12-10 20:23:16,681 - stpipe.Detector1Pipeline.dark_current - INFO - DarkCurrentStep instance created.
2024-12-10 20:23:16,684 - stpipe.Detector1Pipeline.reset - INFO - ResetStep instance created.
2024-12-10 20:23:16,684 - stpipe.Detector1Pipeline.persistence - INFO - PersistenceStep instance created.
2024-12-10 20:23:16,685 - stpipe.Detector1Pipeline.charge_migration - INFO - ChargeMigrationStep instance created.
2024-12-10 20:23:16,687 - stpipe.Detector1Pipeline.jump - INFO - JumpStep instance created.
2024-12-10 20:23:16,688 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - CleanFlickerNoiseStep instance created.
2024-12-10 20:23:16,689 - stpipe.Detector1Pipeline.ramp_fit - INFO - RampFitStep instance created.
2024-12-10 20:23:16,690 - stpipe.Detector1Pipeline.gain_scale - INFO - GainScaleStep instance created.
2024-12-10 20:23:16,872 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline running with args ('1475_f150w/jw01475006001_02201_00017_nis_uncal.fits',).
2024-12-10 20:23:16,895 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: detector1/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_calibrated_ramp: False
steps:
group_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dq_init:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
emicorr:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_intermediate_results: False
user_supplied_reffile: None
nints_to_phase: None
nbins: None
scale_reference: True
onthefly_corr_freq: None
use_n_cycles: 3
saturation:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
n_pix_grow_sat: 1
use_readpatt: True
ipc:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
superbias:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
refpix:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
odd_even_columns: True
use_side_ref_pixels: True
side_smoothing_length: 11
side_gain: 1.0
odd_even_rows: True
ovr_corr_mitigation_ftr: 3.0
preserve_irs2_refpix: False
irs2_mean_subtraction: False
rscd:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
type: baseline
firstframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
lastframe:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
linearity:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_current:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
dark_output: None
average_dark_current: None
reset:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
persistence:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
input_trapsfilled: ''
flag_pers_cutoff: 40.0
save_persistence: False
save_trapsfilled: True
modify_input: False
charge_migration:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
signal_threshold: 21864.0
jump:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
rejection_threshold: 8.0
three_group_rejection_threshold: 6.0
four_group_rejection_threshold: 5.0
maximum_cores: '1'
flag_4_neighbors: False
max_jump_to_flag_neighbors: 200.0
min_jump_to_flag_neighbors: 10.0
after_jump_flag_dn1: 1000
after_jump_flag_time1: 90
after_jump_flag_dn2: 0
after_jump_flag_time2: 0
expand_large_events: True
min_sat_area: 5
min_jump_area: 15.0
expand_factor: 1.75
use_ellipses: False
sat_required_snowball: True
min_sat_radius_extend: 5.0
sat_expand: 0
edge_size: 20
mask_snowball_core_next_int: True
snowball_time_masked_next_int: 4000
find_showers: False
extend_snr_threshold: 1.2
extend_min_area: 90
extend_inner_radius: 1.0
extend_outer_radius: 2.6
extend_ellipse_expand_ratio: 1.1
time_masked_after_shower: 15.0
min_diffs_single_pass: 10
max_extended_radius: 100
minimum_groups: 3
minimum_sigclip_groups: 100
only_use_ints: True
clean_flicker_noise:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
fit_method: median
fit_by_channel: False
background_method: median
background_box_size: None
mask_science_regions: False
n_sigma: 2.0
fit_histogram: False
single_mask: True
user_mask: None
save_mask: False
save_background: False
save_noise: False
ramp_fit:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
algorithm: OLS_C
int_name: ''
save_opt: False
opt_name: ''
suppress_one_group: True
maximum_cores: '1'
gain_scale:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
2024-12-10 20:23:17,062 - stpipe.Detector1Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00017_nis_uncal.fits' reftypes = ['dark', 'gain', 'linearity', 'mask', 'readnoise', 'refpix', 'reset', 'rscd', 'saturation', 'superbias']
2024-12-10 20:23:17,065 - stpipe.Detector1Pipeline - INFO - Prefetch for DARK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits'.
2024-12-10 20:23:17,066 - stpipe.Detector1Pipeline - INFO - Prefetch for GAIN reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits'.
2024-12-10 20:23:17,067 - stpipe.Detector1Pipeline - INFO - Prefetch for LINEARITY reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits'.
2024-12-10 20:23:17,067 - stpipe.Detector1Pipeline - INFO - Prefetch for MASK reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits'.
2024-12-10 20:23:17,068 - stpipe.Detector1Pipeline - INFO - Prefetch for READNOISE reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits'.
2024-12-10 20:23:17,068 - stpipe.Detector1Pipeline - INFO - Prefetch for REFPIX reference file is 'N/A'.
2024-12-10 20:23:17,069 - stpipe.Detector1Pipeline - INFO - Prefetch for RESET reference file is 'N/A'.
2024-12-10 20:23:17,069 - stpipe.Detector1Pipeline - INFO - Prefetch for RSCD reference file is 'N/A'.
2024-12-10 20:23:17,069 - stpipe.Detector1Pipeline - INFO - Prefetch for SATURATION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits'.
2024-12-10 20:23:17,070 - stpipe.Detector1Pipeline - INFO - Prefetch for SUPERBIAS reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits'.
2024-12-10 20:23:17,072 - stpipe.Detector1Pipeline - INFO - Starting calwebb_detector1 ...
2024-12-10 20:23:17,465 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:17,475 - stpipe.Detector1Pipeline.group_scale - INFO - NFRAMES and FRMDIVSR are equal; correction not needed
2024-12-10 20:23:17,476 - stpipe.Detector1Pipeline.group_scale - INFO - Step will be skipped
2024-12-10 20:23:17,477 - stpipe.Detector1Pipeline.group_scale - INFO - Step group_scale done
2024-12-10 20:23:17,657 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:17,677 - stpipe.Detector1Pipeline.dq_init - INFO - Using MASK reference file crds_cache/references/jwst/niriss/jwst_niriss_mask_0017.fits
2024-12-10 20:23:17,902 - stpipe.Detector1Pipeline.dq_init - INFO - Step dq_init done
2024-12-10 20:23:18,084 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:18,103 - stpipe.Detector1Pipeline.saturation - INFO - Using SATURATION reference file crds_cache/references/jwst/niriss/jwst_niriss_saturation_0015.fits
2024-12-10 20:23:18,128 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:18,128 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:18,132 - stpipe.Detector1Pipeline.saturation - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:18,217 - stpipe.Detector1Pipeline.saturation - INFO - Using read_pattern with nframes 4
2024-12-10 20:23:21,411 - stpipe.Detector1Pipeline.saturation - INFO - Detected 4745 saturated pixels
2024-12-10 20:23:21,441 - stpipe.Detector1Pipeline.saturation - INFO - Detected 2 A/D floor pixels
2024-12-10 20:23:21,447 - stpipe.Detector1Pipeline.saturation - INFO - Step saturation done
2024-12-10 20:23:21,629 - stpipe.Detector1Pipeline.ipc - INFO - Step ipc running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:21,629 - stpipe.Detector1Pipeline.ipc - INFO - Step skipped.
2024-12-10 20:23:21,809 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:21,828 - stpipe.Detector1Pipeline.superbias - INFO - Using SUPERBIAS reference file crds_cache/references/jwst/niriss/jwst_niriss_superbias_0183.fits
2024-12-10 20:23:22,099 - stpipe.Detector1Pipeline.superbias - INFO - Step superbias done
2024-12-10 20:23:22,281 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:22,374 - stpipe.Detector1Pipeline.refpix - INFO - NIR full frame data
2024-12-10 20:23:22,374 - stpipe.Detector1Pipeline.refpix - INFO - The following parameters are valid for this mode:
2024-12-10 20:23:22,375 - stpipe.Detector1Pipeline.refpix - INFO - use_side_ref_pixels = True
2024-12-10 20:23:22,375 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_columns = True
2024-12-10 20:23:22,375 - stpipe.Detector1Pipeline.refpix - INFO - side_smoothing_length = 11
2024-12-10 20:23:22,376 - stpipe.Detector1Pipeline.refpix - INFO - side_gain = 1.0
2024-12-10 20:23:22,376 - stpipe.Detector1Pipeline.refpix - INFO - The following parameter is not applicable and is ignored:
2024-12-10 20:23:22,377 - stpipe.Detector1Pipeline.refpix - INFO - odd_even_rows = False
2024-12-10 20:23:26,700 - stpipe.Detector1Pipeline.refpix - INFO - Step refpix done
2024-12-10 20:23:26,883 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:26,903 - stpipe.Detector1Pipeline.linearity - INFO - Using Linearity reference file crds_cache/references/jwst/niriss/jwst_niriss_linearity_0017.fits
2024-12-10 20:23:26,955 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWILLUM does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:26,956 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword LOWRESP does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:26,960 - stpipe.Detector1Pipeline.linearity - WARNING - Keyword UNCERTAIN does not correspond to an existing DQ mnemonic, so will be ignored
2024-12-10 20:23:27,525 - stpipe.Detector1Pipeline.linearity - INFO - Step linearity done
2024-12-10 20:23:27,708 - stpipe.Detector1Pipeline.persistence - INFO - Step persistence running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:27,709 - stpipe.Detector1Pipeline.persistence - INFO - Step skipped.
2024-12-10 20:23:27,890 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:27,910 - stpipe.Detector1Pipeline.dark_current - INFO - Using DARK reference file crds_cache/references/jwst/niriss/jwst_niriss_dark_0169.fits
2024-12-10 20:23:28,176 - stpipe.Detector1Pipeline.dark_current - INFO - Science data nints=1, ngroups=16, nframes=4, groupgap=0
2024-12-10 20:23:28,177 - stpipe.Detector1Pipeline.dark_current - INFO - Dark data nints=1, ngroups=30, nframes=4, groupgap=0
2024-12-10 20:23:28,358 - stpipe.Detector1Pipeline.dark_current - INFO - Step dark_current done
2024-12-10 20:23:28,540 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:28,631 - stpipe.Detector1Pipeline.charge_migration - INFO - Using signal_threshold: 21864.00
2024-12-10 20:23:29,413 - stpipe.Detector1Pipeline.charge_migration - INFO - Step charge_migration done
2024-12-10 20:23:29,594 - stpipe.Detector1Pipeline.jump - INFO - Step jump running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:29,685 - stpipe.Detector1Pipeline.jump - INFO - CR rejection threshold = 8 sigma
2024-12-10 20:23:29,686 - stpipe.Detector1Pipeline.jump - INFO - Maximum cores to use = 1
2024-12-10 20:23:29,697 - stpipe.Detector1Pipeline.jump - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:23:29,720 - stpipe.Detector1Pipeline.jump - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:23:29,917 - stpipe.Detector1Pipeline.jump - INFO - Executing two-point difference method
2024-12-10 20:23:40,132 - stpipe.Detector1Pipeline.jump - INFO - Flagging Snowballs
2024-12-10 20:23:41,601 - stpipe.Detector1Pipeline.jump - INFO - Total snowballs = 41
2024-12-10 20:23:41,602 - stpipe.Detector1Pipeline.jump - INFO - Total elapsed time = 11.6842 sec
2024-12-10 20:23:41,688 - stpipe.Detector1Pipeline.jump - INFO - The execution time in seconds: 12.084302
2024-12-10 20:23:41,691 - stpipe.Detector1Pipeline.jump - INFO - Step jump done
2024-12-10 20:23:41,865 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step clean_flicker_noise running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:41,865 - stpipe.Detector1Pipeline.clean_flicker_noise - INFO - Step skipped.
2024-12-10 20:23:42,041 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit running with args (<RampModel(1, 16, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:42,154 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using READNOISE reference file: crds_cache/references/jwst/niriss/jwst_niriss_readnoise_0005.fits
2024-12-10 20:23:42,154 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using GAIN reference file: crds_cache/references/jwst/niriss/jwst_niriss_gain_0006.fits
2024-12-10 20:23:42,182 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using algorithm = OLS_C
2024-12-10 20:23:42,183 - stpipe.Detector1Pipeline.ramp_fit - INFO - Using weighting = optimal
2024-12-10 20:23:42,746 - stpipe.Detector1Pipeline.ramp_fit - INFO - Number of multiprocessing slices: 1
2024-12-10 20:23:48,375 - stpipe.Detector1Pipeline.ramp_fit - INFO - Ramp Fitting C Time: 5.624956846237183
2024-12-10 20:23:48,429 - stpipe.Detector1Pipeline.ramp_fit - INFO - Step ramp_fit done
2024-12-10 20:23:48,612 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<ImageModel(2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:48,641 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:23:48,641 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:23:48,644 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:23:48,827 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale running with args (<CubeModel(1, 2048, 2048) from jw01475006001_02201_00017_nis_uncal.fits>,).
2024-12-10 20:23:48,858 - stpipe.Detector1Pipeline.gain_scale - INFO - GAINFACT not found in gain reference file
2024-12-10 20:23:48,858 - stpipe.Detector1Pipeline.gain_scale - INFO - Step will be skipped
2024-12-10 20:23:48,860 - stpipe.Detector1Pipeline.gain_scale - INFO - Step gain_scale done
2024-12-10 20:23:49,012 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00017_nis_rateints.fits
2024-12-10 20:23:49,013 - stpipe.Detector1Pipeline - INFO - ... ending calwebb_detector1
2024-12-10 20:23:49,015 - stpipe.Detector1Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:23:49,161 - stpipe.Detector1Pipeline - INFO - Saved model in detector1/jw01475006001_02201_00017_nis_rate.fits
2024-12-10 20:23:49,162 - stpipe.Detector1Pipeline - INFO - Step Detector1Pipeline done
2024-12-10 20:23:49,162 - stpipe - INFO - Results used jwst version: 1.16.1
Identify *_rate.fits files#
rate_files = sorted(glob.glob(os.path.join(det1_dir, '*_rate.fits')))
Verify which pipeline steps were run and which calibration reference files were applied#
The header contains information about which calibration steps were completed and skipped and which reference files were used to process the data.
# Read in file as datamodel
rate_f = datamodels.open(rate_files[0])
rate_f.meta.cal_step.instance
{'charge_migration': 'COMPLETE',
'clean_flicker_noise': 'SKIPPED',
'dark_sub': 'COMPLETE',
'dq_init': 'COMPLETE',
'gain_scale': 'SKIPPED',
'group_scale': 'SKIPPED',
'ipc': 'SKIPPED',
'jump': 'COMPLETE',
'linearity': 'COMPLETE',
'persistence': 'SKIPPED',
'ramp_fit': 'COMPLETE',
'refpix': 'COMPLETE',
'saturation': 'COMPLETE',
'superbias': 'COMPLETE'}
Check which reference files were used to calibrate the dataset:
rate_f.meta.ref_file.instance
{'crds': {'context_used': 'jwst_1303.pmap', 'sw_version': '12.0.8'},
'dark': {'name': 'crds://jwst_niriss_dark_0169.fits'},
'gain': {'name': 'crds://jwst_niriss_gain_0006.fits'},
'linearity': {'name': 'crds://jwst_niriss_linearity_0017.fits'},
'mask': {'name': 'crds://jwst_niriss_mask_0017.fits'},
'readnoise': {'name': 'crds://jwst_niriss_readnoise_0005.fits'},
'saturation': {'name': 'crds://jwst_niriss_saturation_0015.fits'},
'superbias': {'name': 'crds://jwst_niriss_superbias_0183.fits'}}
Display rate image#
Visualize a countrate image, using the dataset from the first dither position as an example.
# Create an Imviz instance and set up default viewer
imviz_rate = Imviz()
viewer_rate = imviz_rate.default_viewer
# Read in the science array for our visualization dataset:
rate_science = rate_f.data
# Load the dataset into Imviz
imviz_rate.load_data(rate_science)
# Visualize the dataset:
imviz_rate.show()

plotopt = imviz_rate.plugins['Plot Options']
plotopt.stretch_function = 'sqrt'
plotopt.image_colormap = 'Viridis'
plotopt.stretch_preset = '95%'
plotopt.zoom_radius = 1024
The viewer looks like this:
viewer_rate.save('./rate_science.png')
Image('./rate_science.png')
3. Stage 2 Processing #
In the Image2 stage of the pipeline, calibrated unrectified data products are created (*_cal.fits or *_calints.fits files, depending on whether the input files are *_rate.fits or *_rateints.fits).
In this pipeline processing stage, the world coordinate system (WCS) is assigned, the data are flat fielded, and a photometric calibration is applied to convert from units of countrate (ADU/s) to surface brightness (MJy/sr).
By default, the background subtraction step and the resampling step are turned off for NIRISS at this stage of the pipeline. The background subtraction is turned off since there is no background template for the imaging mode and the local background is removed during the background correction for photometric measurements around individual sources. The resampling step occurs during the Image3 stage by default. While the resampling step can be turned on during the Image2 stage to, e.g., generate a source catalog for each image, the data quality from the Image3 stage will be better since the bad pixels, which adversely affect both the centroids and photometry in individual images, will be mostly removed.
# Define directory to save output from Image2
image2_dir = 'image2/'
# Create directory if it does not exist
if not os.path.isdir(image2_dir):
os.mkdir(image2_dir)
# Run Image2 stage of pipeline, specifying:
# output directory to save *_cal.fits files
# save_results flag set to True so the rate files are saved
for rate in rate_files:
cal_result = Image2Pipeline.call(rate,
output_dir=image2_dir,
save_results=True)
2024-12-10 20:23:50,129 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf 1.2 K bytes (1 / 1 files) (0 / 1.2 K bytes)
2024-12-10 20:23:50,282 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:23:50,292 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf 1.3 K bytes (1 / 1 files) (0 / 1.3 K bytes)
2024-12-10 20:23:50,427 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:23:50,440 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:23:50,441 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:23:50,442 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:23:50,443 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:23:50,444 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:23:50,446 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:23:50,637 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00001_nis_rate.fits',).
2024-12-10 20:23:50,646 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:23:50,698 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00001_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:23:50,701 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits 16.8 M bytes (1 / 5 files) (0 / 83.9 M bytes)
2024-12-10 20:23:51,987 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf 9.9 K bytes (2 / 5 files) (16.8 M / 83.9 M bytes)
2024-12-10 20:23:52,102 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf 5.4 K bytes (3 / 5 files) (16.8 M / 83.9 M bytes)
2024-12-10 20:23:52,240 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits 67.1 M bytes (4 / 5 files) (16.8 M / 83.9 M bytes)
2024-12-10 20:23:55,416 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits 11.5 K bytes (5 / 5 files) (83.9 M / 83.9 M bytes)
2024-12-10 20:23:55,539 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:23:55,539 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:23:55,540 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:23:55,540 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:23:55,541 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:23:55,541 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:23:55,542 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:23:55,542 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:23:55,542 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:23:55,543 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:23:55,544 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:23:55,544 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:23:55,544 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:23:55,545 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:23:55,545 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:23:55,546 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:23:55,546 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:23:55,547 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:23:55,548 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:23:55,548 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:23:55,548 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:23:55,549 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:23:55,550 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00001_nis
2024-12-10 20:23:55,550 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00001_nis_rate.fits ...
2024-12-10 20:23:55,794 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00001_nis_image2pipeline.fits>,).
2024-12-10 20:23:55,990 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:23:56,054 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.741854239 -26.792479051 303.782502890 -26.783393014 303.772199160 -26.747414924 303.731588611 -26.756482660
2024-12-10 20:23:56,055 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.741854239 -26.792479051 303.782502890 -26.783393014 303.772199160 -26.747414924 303.731588611 -26.756482660
2024-12-10 20:23:56,056 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:23:56,059 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:23:56,061 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:23:56,124 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:23:56,322 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00001_nis_image2pipeline.fits>,).
2024-12-10 20:23:56,415 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:23:56,416 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:23:56,417 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:23:56,417 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:23:56,585 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:23:56,784 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00001_nis_image2pipeline.fits>,).
2024-12-10 20:23:56,808 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:23:56,809 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:23:56,855 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:23:56,855 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:23:56,856 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:23:56,856 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:23:56,857 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:23:56,889 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:23:56,890 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:23:56,891 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:23:56,934 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:23:57,133 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00001_nis_image2pipeline.fits>,).
2024-12-10 20:23:57,133 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:23:57,135 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00001_nis
2024-12-10 20:23:57,135 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:23:57,136 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:23:57,408 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00001_nis_cal.fits
2024-12-10 20:23:57,408 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:23:57,409 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:23:57,463 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:23:57,473 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:23:57,485 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:23:57,487 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:23:57,488 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:23:57,489 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:23:57,490 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:23:57,492 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:23:57,694 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00002_nis_rate.fits',).
2024-12-10 20:23:57,703 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:23:57,758 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00002_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:23:57,761 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:23:57,762 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:23:57,762 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:23:57,763 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:23:57,763 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:23:57,763 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:23:57,764 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:23:57,765 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:23:57,765 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:23:57,766 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:23:57,766 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:23:57,766 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:23:57,767 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:23:57,767 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:23:57,768 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:23:57,768 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:23:57,768 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:23:57,769 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:23:57,770 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:23:57,770 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:23:57,770 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:23:57,772 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:23:57,772 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00002_nis
2024-12-10 20:23:57,773 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00002_nis_rate.fits ...
2024-12-10 20:23:58,026 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00002_nis_image2pipeline.fits>,).
2024-12-10 20:23:58,212 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:23:58,272 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.738214137 -26.798033307 303.778865034 -26.788948185 303.768561815 -26.752969863 303.727949025 -26.762036684
2024-12-10 20:23:58,273 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.738214137 -26.798033307 303.778865034 -26.788948185 303.768561815 -26.752969863 303.727949025 -26.762036684
2024-12-10 20:23:58,274 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:23:58,276 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:23:58,277 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:23:58,329 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:23:58,533 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00002_nis_image2pipeline.fits>,).
2024-12-10 20:23:58,622 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:23:58,622 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:23:58,623 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:23:58,624 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:23:58,792 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:23:58,996 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00002_nis_image2pipeline.fits>,).
2024-12-10 20:23:59,021 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:23:59,022 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:23:59,065 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:23:59,065 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:23:59,066 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:23:59,066 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:23:59,067 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:23:59,095 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:23:59,095 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:23:59,097 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:23:59,139 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:23:59,342 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00002_nis_image2pipeline.fits>,).
2024-12-10 20:23:59,343 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:23:59,345 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00002_nis
2024-12-10 20:23:59,346 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:23:59,346 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:23:59,617 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00002_nis_cal.fits
2024-12-10 20:23:59,618 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:23:59,618 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:23:59,672 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:23:59,682 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:23:59,694 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:23:59,695 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:23:59,697 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:23:59,698 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:23:59,698 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:23:59,700 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:23:59,895 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00003_nis_rate.fits',).
2024-12-10 20:23:59,904 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:23:59,958 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00003_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:23:59,962 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:23:59,962 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:23:59,963 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:23:59,963 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:23:59,964 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:23:59,965 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:23:59,965 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:23:59,966 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:23:59,966 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:23:59,967 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:23:59,967 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:23:59,967 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:23:59,968 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:23:59,968 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:23:59,969 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:23:59,969 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:23:59,969 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:23:59,970 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:23:59,970 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:23:59,971 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:23:59,971 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:23:59,972 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:23:59,973 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00003_nis
2024-12-10 20:23:59,973 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00003_nis_rate.fits ...
2024-12-10 20:24:00,223 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00003_nis_image2pipeline.fits>,).
2024-12-10 20:24:00,409 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:00,469 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.748076235 -26.795728135 303.788725488 -26.786640096 303.778419241 -26.750662514 303.737808091 -26.759732249
2024-12-10 20:24:00,470 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.748076235 -26.795728135 303.788725488 -26.786640096 303.778419241 -26.750662514 303.737808091 -26.759732249
2024-12-10 20:24:00,471 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:00,473 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:00,474 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:00,526 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:00,736 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00003_nis_image2pipeline.fits>,).
2024-12-10 20:24:00,824 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:00,824 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:00,825 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:00,825 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:00,979 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:01,184 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00003_nis_image2pipeline.fits>,).
2024-12-10 20:24:01,209 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:01,210 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:01,256 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:01,256 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:01,257 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:01,257 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:01,258 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:01,286 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:01,286 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:01,288 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:01,330 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:01,538 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00003_nis_image2pipeline.fits>,).
2024-12-10 20:24:01,539 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:01,540 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00003_nis
2024-12-10 20:24:01,541 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:01,542 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:01,814 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00003_nis_cal.fits
2024-12-10 20:24:01,815 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:01,815 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:01,872 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:01,882 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:01,894 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:01,896 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:01,897 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:01,898 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:01,899 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:01,901 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:02,100 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00004_nis_rate.fits',).
2024-12-10 20:24:02,108 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:02,164 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00004_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:02,168 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:02,168 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:02,169 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:02,169 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:02,170 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:02,170 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:02,171 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:02,171 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:02,172 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:02,172 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:02,173 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:02,173 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:02,174 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:02,174 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:02,175 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:02,175 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:02,175 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:02,177 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:02,177 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:02,178 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:02,178 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:02,179 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:02,180 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00004_nis
2024-12-10 20:24:02,180 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00004_nis_rate.fits ...
2024-12-10 20:24:02,433 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00004_nis_image2pipeline.fits>,).
2024-12-10 20:24:02,622 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:02,681 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.745493505 -26.786924996 303.786139823 -26.777837735 303.775835240 -26.741859955 303.735227018 -26.750928915
2024-12-10 20:24:02,682 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.745493505 -26.786924996 303.786139823 -26.777837735 303.775835240 -26.741859955 303.735227018 -26.750928915
2024-12-10 20:24:02,683 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:02,685 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:02,686 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:02,738 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:02,941 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00004_nis_image2pipeline.fits>,).
2024-12-10 20:24:03,030 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:03,031 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:03,031 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:03,032 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:03,198 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:03,401 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00004_nis_image2pipeline.fits>,).
2024-12-10 20:24:03,426 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:03,426 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:03,473 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:03,473 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:03,474 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:03,474 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:03,475 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:03,502 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:03,503 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:03,504 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:03,546 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:03,748 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00004_nis_image2pipeline.fits>,).
2024-12-10 20:24:03,749 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:03,750 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00004_nis
2024-12-10 20:24:03,751 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:03,751 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:04,025 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00004_nis_cal.fits
2024-12-10 20:24:04,025 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:04,026 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:04,082 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:04,091 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:04,104 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:04,105 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:04,107 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:04,108 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:04,109 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:04,110 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:04,316 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00005_nis_rate.fits',).
2024-12-10 20:24:04,324 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:04,381 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00005_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:04,384 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:04,385 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:04,385 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:04,385 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:04,386 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:04,386 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:04,387 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:04,387 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:04,388 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:04,389 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:04,389 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:04,389 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:04,390 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:04,390 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:04,390 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:04,391 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:04,391 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:04,392 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:04,393 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:04,393 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:04,393 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:04,394 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:04,395 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00005_nis
2024-12-10 20:24:04,395 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00005_nis_rate.fits ...
2024-12-10 20:24:04,650 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00005_nis_image2pipeline.fits>,).
2024-12-10 20:24:04,837 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:04,898 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.735631928 -26.789230123 303.776279861 -26.780145676 303.765978192 -26.744167183 303.725368360 -26.753233330
2024-12-10 20:24:04,898 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.735631928 -26.789230123 303.776279861 -26.780145676 303.765978192 -26.744167183 303.725368360 -26.753233330
2024-12-10 20:24:04,899 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:04,901 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:04,902 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:04,958 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:05,163 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00005_nis_image2pipeline.fits>,).
2024-12-10 20:24:05,253 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:05,254 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:05,254 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:05,255 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:05,412 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:05,617 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00005_nis_image2pipeline.fits>,).
2024-12-10 20:24:05,642 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:05,642 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:05,688 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:05,688 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:05,689 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:05,689 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:05,690 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:05,717 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:05,718 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:05,720 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:05,761 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:05,964 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00005_nis_image2pipeline.fits>,).
2024-12-10 20:24:05,965 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:05,966 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00005_nis
2024-12-10 20:24:05,967 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:05,968 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:06,237 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00005_nis_cal.fits
2024-12-10 20:24:06,238 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:06,238 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:06,293 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:06,302 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:06,314 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:06,315 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:06,317 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:06,318 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:06,319 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:06,320 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:06,519 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00006_nis_rate.fits',).
2024-12-10 20:24:06,527 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:06,582 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00006_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:06,586 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:06,587 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:06,587 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:06,588 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:06,588 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:06,588 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:06,589 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:06,589 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:06,591 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:06,591 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:06,591 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:06,592 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:06,592 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:06,593 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:06,593 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:06,593 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:06,594 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:06,594 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:06,595 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:06,595 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:06,595 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:06,596 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:06,597 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00006_nis
2024-12-10 20:24:06,597 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00006_nis_rate.fits ...
2024-12-10 20:24:06,849 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00006_nis_image2pipeline.fits>,).
2024-12-10 20:24:07,033 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:07,092 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.736922794 -26.793631848 303.777572181 -26.784546967 303.767269630 -26.748568584 303.726658346 -26.757635166
2024-12-10 20:24:07,093 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.736922794 -26.793631848 303.777572181 -26.784546967 303.767269630 -26.748568584 303.726658346 -26.757635166
2024-12-10 20:24:07,094 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:07,096 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:07,096 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:07,149 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:07,356 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00006_nis_image2pipeline.fits>,).
2024-12-10 20:24:07,442 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:07,442 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:07,443 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:07,443 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:07,604 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:07,810 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00006_nis_image2pipeline.fits>,).
2024-12-10 20:24:07,834 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:07,835 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:07,879 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:07,880 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:07,880 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:07,881 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:07,881 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:07,909 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:07,910 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:07,911 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:07,953 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:08,154 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00006_nis_image2pipeline.fits>,).
2024-12-10 20:24:08,155 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:08,156 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00006_nis
2024-12-10 20:24:08,157 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:08,157 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:08,427 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00006_nis_cal.fits
2024-12-10 20:24:08,428 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:08,428 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:08,482 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:08,492 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:08,505 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:08,506 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:08,507 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:08,508 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:08,509 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:08,511 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:08,710 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00007_nis_rate.fits',).
2024-12-10 20:24:08,720 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:08,775 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00007_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:08,778 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:08,779 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:08,780 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:08,780 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:08,781 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:08,781 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:08,782 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:08,782 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:08,783 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:08,784 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:08,784 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:08,785 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:08,785 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:08,786 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:08,786 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:08,786 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:08,787 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:08,787 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:08,788 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:08,788 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:08,789 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:08,790 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:08,790 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00007_nis
2024-12-10 20:24:08,791 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00007_nis_rate.fits ...
2024-12-10 20:24:09,048 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00007_nis_image2pipeline.fits>,).
2024-12-10 20:24:09,233 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:09,292 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.743144738 -26.796881330 303.783794840 -26.787794847 303.773490215 -26.751816871 303.732878218 -26.760885052
2024-12-10 20:24:09,293 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.743144738 -26.796881330 303.783794840 -26.787794847 303.773490215 -26.751816871 303.732878218 -26.760885052
2024-12-10 20:24:09,293 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:09,296 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:09,296 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:09,350 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:09,551 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00007_nis_image2pipeline.fits>,).
2024-12-10 20:24:09,649 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:09,650 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:09,650 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:09,651 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:09,806 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:10,013 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00007_nis_image2pipeline.fits>,).
2024-12-10 20:24:10,039 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:10,039 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:10,085 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:10,086 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:10,086 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:10,087 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:10,087 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:10,115 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:10,115 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:10,117 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:10,158 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:10,363 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00007_nis_image2pipeline.fits>,).
2024-12-10 20:24:10,364 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:10,365 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00007_nis
2024-12-10 20:24:10,366 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:10,367 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:10,637 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00007_nis_cal.fits
2024-12-10 20:24:10,638 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:10,639 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:10,695 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:10,704 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:10,717 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:10,718 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:10,719 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:10,721 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:10,722 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:10,723 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:10,925 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00008_nis_rate.fits',).
2024-12-10 20:24:10,934 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:10,990 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00008_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:10,994 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:10,994 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:10,995 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:10,995 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:10,996 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:10,996 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:10,996 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:10,997 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:10,998 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:10,998 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:10,999 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:10,999 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:10,999 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:11,000 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:11,000 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:11,000 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:11,001 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:11,001 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:11,001 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:11,002 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:11,002 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:11,003 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:11,004 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00008_nis
2024-12-10 20:24:11,004 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00008_nis_rate.fits ...
2024-12-10 20:24:11,250 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00008_nis_image2pipeline.fits>,).
2024-12-10 20:24:11,432 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:11,491 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.746784872 -26.791326555 303.787432662 -26.782238924 303.777127267 -26.746261238 303.736517577 -26.755330567
2024-12-10 20:24:11,491 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.746784872 -26.791326555 303.787432662 -26.782238924 303.777127267 -26.746261238 303.736517577 -26.755330567
2024-12-10 20:24:11,492 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:11,495 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:11,495 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:11,546 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:11,745 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00008_nis_image2pipeline.fits>,).
2024-12-10 20:24:11,830 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:11,831 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:11,831 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:11,832 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:11,990 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:12,197 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00008_nis_image2pipeline.fits>,).
2024-12-10 20:24:12,222 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:12,222 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:12,268 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:12,269 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:12,269 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:12,270 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:12,270 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:12,298 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:12,299 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:12,300 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:12,343 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:12,542 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00008_nis_image2pipeline.fits>,).
2024-12-10 20:24:12,543 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:12,545 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00008_nis
2024-12-10 20:24:12,545 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:12,546 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:12,816 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00008_nis_cal.fits
2024-12-10 20:24:12,816 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:12,817 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:12,872 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:12,881 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:12,894 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:12,895 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:12,896 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:12,897 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:12,898 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:12,899 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:13,101 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00009_nis_rate.fits',).
2024-12-10 20:24:13,109 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:13,164 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00009_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:13,167 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:13,168 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:13,168 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:13,168 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:13,169 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:13,169 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:13,170 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:13,170 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:13,171 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:13,172 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:13,172 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:13,172 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:13,173 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:13,173 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:13,174 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:13,174 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:13,174 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:13,175 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:13,175 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:13,176 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:13,176 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:13,177 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:13,179 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00009_nis
2024-12-10 20:24:13,180 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00009_nis_rate.fits ...
2024-12-10 20:24:13,431 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00009_nis_image2pipeline.fits>,).
2024-12-10 20:24:13,614 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:13,674 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.740562612 -26.788077734 303.781209713 -26.778991797 303.770906496 -26.743013682 303.730297492 -26.752081318
2024-12-10 20:24:13,674 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.740562612 -26.788077734 303.781209713 -26.778991797 303.770906496 -26.743013682 303.730297492 -26.752081318
2024-12-10 20:24:13,675 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:13,677 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:13,677 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:13,736 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:13,937 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00009_nis_image2pipeline.fits>,).
2024-12-10 20:24:14,024 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:14,024 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:14,025 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:14,025 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:14,183 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:14,386 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00009_nis_image2pipeline.fits>,).
2024-12-10 20:24:14,410 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:14,411 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:14,456 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:14,457 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:14,457 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:14,458 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:14,458 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:14,486 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:14,486 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:14,488 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:14,529 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:14,730 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00009_nis_image2pipeline.fits>,).
2024-12-10 20:24:14,731 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:14,732 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00009_nis
2024-12-10 20:24:14,733 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:14,734 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:15,006 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00009_nis_cal.fits
2024-12-10 20:24:15,007 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:15,007 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:15,066 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:15,075 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:15,088 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:15,089 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:15,091 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:15,092 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:15,093 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:15,095 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:15,295 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00010_nis_rate.fits',).
2024-12-10 20:24:15,304 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:15,358 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00010_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:15,361 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:15,362 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:15,363 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:15,363 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:15,363 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:15,364 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:15,365 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:15,366 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:15,366 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:15,367 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:15,367 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:15,367 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:15,368 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:15,368 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:15,369 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:15,369 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:15,369 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:15,371 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:15,371 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:15,372 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:15,372 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:15,373 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:15,373 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00010_nis
2024-12-10 20:24:15,374 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00010_nis_rate.fits ...
2024-12-10 20:24:15,625 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00010_nis_image2pipeline.fits>,).
2024-12-10 20:24:15,811 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:15,870 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.738742913 -26.790854726 303.779391177 -26.781769388 303.769088372 -26.745791122 303.728478209 -26.754858159
2024-12-10 20:24:15,871 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.738742913 -26.790854726 303.779391177 -26.781769388 303.769088372 -26.745791122 303.728478209 -26.754858159
2024-12-10 20:24:15,872 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:15,874 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:15,875 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:15,926 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:16,129 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00010_nis_image2pipeline.fits>,).
2024-12-10 20:24:16,212 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:16,213 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:16,213 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:16,214 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:16,367 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:16,570 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00010_nis_image2pipeline.fits>,).
2024-12-10 20:24:16,595 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:16,595 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:16,641 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:16,641 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:16,642 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:16,642 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:16,642 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:16,670 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:16,671 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:16,672 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:16,714 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:16,916 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00010_nis_image2pipeline.fits>,).
2024-12-10 20:24:16,916 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:16,918 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00010_nis
2024-12-10 20:24:16,919 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:16,919 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:17,192 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00010_nis_cal.fits
2024-12-10 20:24:17,193 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:17,193 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:17,251 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:17,262 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:17,275 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:17,276 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:17,277 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:17,278 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:17,279 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:17,281 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:17,486 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00011_nis_rate.fits',).
2024-12-10 20:24:17,494 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:17,549 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00011_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:17,552 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:17,553 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:17,553 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:17,554 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:17,554 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:17,554 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:17,555 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:17,555 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:17,556 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:17,557 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:17,558 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:17,558 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:17,558 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:17,559 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:17,559 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:17,559 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:17,560 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:17,561 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:17,561 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:17,561 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:17,562 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:17,563 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:17,563 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00011_nis
2024-12-10 20:24:17,563 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00011_nis_rate.fits ...
2024-12-10 20:24:17,814 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00011_nis_image2pipeline.fits>,).
2024-12-10 20:24:17,996 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:18,056 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.740034198 -26.795256243 303.780683961 -26.786170628 303.770380447 -26.750192432 303.729768789 -26.759259746
2024-12-10 20:24:18,057 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.740034198 -26.795256243 303.780683961 -26.786170628 303.770380447 -26.750192432 303.729768789 -26.759259746
2024-12-10 20:24:18,057 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:18,060 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:18,060 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:18,112 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:18,317 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00011_nis_image2pipeline.fits>,).
2024-12-10 20:24:18,403 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:18,404 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:18,404 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:18,405 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:18,566 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:18,767 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00011_nis_image2pipeline.fits>,).
2024-12-10 20:24:18,792 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:18,793 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:18,838 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:18,839 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:18,839 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:18,840 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:18,840 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:18,868 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:18,869 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:18,870 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:18,911 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:19,114 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00011_nis_image2pipeline.fits>,).
2024-12-10 20:24:19,115 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:19,117 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00011_nis
2024-12-10 20:24:19,118 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:19,118 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:19,389 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00011_nis_cal.fits
2024-12-10 20:24:19,390 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:19,390 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:19,445 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:19,454 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:19,467 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:19,468 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:19,469 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:19,470 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:19,471 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:19,473 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:19,674 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00012_nis_rate.fits',).
2024-12-10 20:24:19,682 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:19,737 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00012_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:19,740 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:19,740 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:19,741 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:19,741 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:19,741 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:19,742 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:19,742 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:19,743 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:19,743 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:19,743 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:19,744 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:19,744 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:19,744 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:19,745 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:19,745 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:19,745 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:19,746 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:19,746 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:19,747 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:19,747 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:19,747 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:19,748 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:19,749 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00012_nis
2024-12-10 20:24:19,749 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00012_nis_rate.fits ...
2024-12-10 20:24:19,995 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00012_nis_image2pipeline.fits>,).
2024-12-10 20:24:20,180 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:20,239 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.744965132 -26.794103704 303.785614069 -26.785016615 303.775309024 -26.749038793 303.734698191 -26.758107579
2024-12-10 20:24:20,240 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.744965132 -26.794103704 303.785614069 -26.785016615 303.775309024 -26.749038793 303.734698191 -26.758107579
2024-12-10 20:24:20,241 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:20,243 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:20,244 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:20,296 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:20,496 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00012_nis_image2pipeline.fits>,).
2024-12-10 20:24:20,582 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:20,583 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:20,583 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:20,584 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:20,734 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:20,934 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00012_nis_image2pipeline.fits>,).
2024-12-10 20:24:20,959 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:20,960 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:21,007 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:21,008 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:21,008 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:21,008 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:21,009 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:21,037 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:21,038 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:21,039 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:21,080 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:21,279 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00012_nis_image2pipeline.fits>,).
2024-12-10 20:24:21,280 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:21,282 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00012_nis
2024-12-10 20:24:21,283 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:21,283 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:21,551 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00012_nis_cal.fits
2024-12-10 20:24:21,552 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:21,553 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:21,615 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:21,625 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:21,637 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:21,638 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:21,639 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:21,640 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:21,641 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:21,643 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:21,846 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00013_nis_rate.fits',).
2024-12-10 20:24:21,854 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:21,914 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00013_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:21,918 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:21,918 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:21,919 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:21,919 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:21,920 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:21,920 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:21,921 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:21,922 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:21,922 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:21,922 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:21,923 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:21,923 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:21,924 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:21,924 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:21,926 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:21,926 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:21,927 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:21,927 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:21,928 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:21,929 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:21,929 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:21,930 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:21,931 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00013_nis
2024-12-10 20:24:21,931 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00013_nis_rate.fits ...
2024-12-10 20:24:22,186 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00013_nis_image2pipeline.fits>,).
2024-12-10 20:24:22,373 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:22,437 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.743673743 -26.789702172 303.784321194 -26.780615409 303.774016911 -26.744637504 303.733407559 -26.753705966
2024-12-10 20:24:22,438 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.743673743 -26.789702172 303.784321194 -26.780615409 303.774016911 -26.744637504 303.733407559 -26.753705966
2024-12-10 20:24:22,438 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:22,441 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:22,441 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:22,493 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:22,694 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00013_nis_image2pipeline.fits>,).
2024-12-10 20:24:22,781 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:22,781 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:22,782 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:22,782 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:22,950 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:23,153 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00013_nis_image2pipeline.fits>,).
2024-12-10 20:24:23,178 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:23,178 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:23,225 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:23,225 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:23,226 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:23,226 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:23,226 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:23,254 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:23,254 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:23,255 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:23,296 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:23,491 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00013_nis_image2pipeline.fits>,).
2024-12-10 20:24:23,492 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:23,493 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00013_nis
2024-12-10 20:24:23,494 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:23,495 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:23,763 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00013_nis_cal.fits
2024-12-10 20:24:23,763 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:23,764 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:23,822 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:23,832 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:23,844 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:23,845 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:23,847 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:23,848 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:23,849 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:23,850 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:24,051 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00014_nis_rate.fits',).
2024-12-10 20:24:24,060 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:24,114 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00014_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:24,118 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:24,118 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:24,119 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:24,119 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:24,120 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:24,120 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:24,122 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:24,122 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:24,123 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:24,124 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:24,124 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:24,124 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:24,125 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:24,125 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:24,126 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:24,126 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:24,127 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:24,128 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:24,128 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:24,128 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:24,129 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:24,130 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:24,130 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00014_nis
2024-12-10 20:24:24,130 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00014_nis_rate.fits ...
2024-12-10 20:24:24,377 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00014_nis_image2pipeline.fits>,).
2024-12-10 20:24:24,560 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:24,619 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.741208296 -26.790278517 303.781856145 -26.781192440 303.771552571 -26.745214361 303.730942822 -26.754282137
2024-12-10 20:24:24,619 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.741208296 -26.790278517 303.781856145 -26.781192440 303.771552571 -26.745214361 303.730942822 -26.754282137
2024-12-10 20:24:24,620 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:24,622 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:24,623 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:24,675 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:24,875 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00014_nis_image2pipeline.fits>,).
2024-12-10 20:24:24,961 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:24,962 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:24,963 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:24,963 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:25,115 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:25,315 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00014_nis_image2pipeline.fits>,).
2024-12-10 20:24:25,340 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:25,341 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:25,387 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:25,388 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:25,388 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:25,389 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:25,389 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:25,417 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:25,418 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:25,419 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:25,460 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:25,661 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00014_nis_image2pipeline.fits>,).
2024-12-10 20:24:25,661 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:25,663 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00014_nis
2024-12-10 20:24:25,664 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:25,664 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:25,933 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00014_nis_cal.fits
2024-12-10 20:24:25,934 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:25,934 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:25,989 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:25,999 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:26,011 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:26,012 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:26,013 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:26,015 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:26,016 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:26,017 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:26,215 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00015_nis_rate.fits',).
2024-12-10 20:24:26,224 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:26,277 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00015_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:26,281 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:26,281 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:26,282 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:26,282 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:26,282 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:26,283 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:26,284 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:26,284 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:26,285 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:26,286 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:26,286 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:26,287 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:26,287 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:26,287 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:26,288 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:26,288 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:26,289 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:26,290 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:26,291 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:26,291 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:26,292 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:26,293 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:26,293 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00015_nis
2024-12-10 20:24:26,294 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00015_nis_rate.fits ...
2024-12-10 20:24:26,542 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00015_nis_image2pipeline.fits>,).
2024-12-10 20:24:26,724 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:26,783 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.739388537 -26.793055529 303.780037545 -26.783970036 303.769734367 -26.747991809 303.729123462 -26.757059002
2024-12-10 20:24:26,784 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.739388537 -26.793055529 303.780037545 -26.783970036 303.769734367 -26.747991809 303.729123462 -26.757059002
2024-12-10 20:24:26,784 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:26,787 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:26,787 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:26,842 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:27,038 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00015_nis_image2pipeline.fits>,).
2024-12-10 20:24:27,124 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:27,125 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:27,125 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:27,126 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:27,278 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:27,483 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00015_nis_image2pipeline.fits>,).
2024-12-10 20:24:27,507 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:27,508 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:27,549 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:27,550 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:27,550 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:27,551 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:27,551 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:27,579 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:27,579 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:27,580 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:27,621 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:27,825 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00015_nis_image2pipeline.fits>,).
2024-12-10 20:24:27,825 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:27,827 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00015_nis
2024-12-10 20:24:27,828 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:27,828 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:28,096 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00015_nis_cal.fits
2024-12-10 20:24:28,096 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:28,097 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:28,151 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:28,161 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:28,173 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:28,174 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:28,175 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:28,176 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:28,177 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:28,179 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:28,378 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00016_nis_rate.fits',).
2024-12-10 20:24:28,386 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:28,440 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00016_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:28,443 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:28,444 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:28,444 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:28,445 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:28,445 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:28,446 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:28,446 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:28,447 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:28,447 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:28,448 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:28,448 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:28,449 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:28,449 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:28,449 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:28,450 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:28,451 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:28,451 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:28,452 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:28,452 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:28,453 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:28,453 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:28,454 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:28,455 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00016_nis
2024-12-10 20:24:28,455 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00016_nis_rate.fits ...
2024-12-10 20:24:28,704 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00016_nis_image2pipeline.fits>,).
2024-12-10 20:24:28,886 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:28,946 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.742499665 -26.794680027 303.783149008 -26.785593654 303.772844705 -26.749615651 303.732233466 -26.758683722
2024-12-10 20:24:28,946 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.742499665 -26.794680027 303.783149008 -26.785593654 303.772844705 -26.749615651 303.732233466 -26.758683722
2024-12-10 20:24:28,947 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:28,949 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:28,949 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:29,001 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:29,205 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00016_nis_image2pipeline.fits>,).
2024-12-10 20:24:29,291 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:29,292 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:29,292 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:29,293 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:29,445 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:29,649 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00016_nis_image2pipeline.fits>,).
2024-12-10 20:24:29,673 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:29,674 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:29,719 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:29,720 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:29,720 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:29,721 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:29,721 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:29,749 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:29,749 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:29,751 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:29,792 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:29,994 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00016_nis_image2pipeline.fits>,).
2024-12-10 20:24:29,995 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:29,997 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00016_nis
2024-12-10 20:24:29,998 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:29,998 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:30,267 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00016_nis_cal.fits
2024-12-10 20:24:30,268 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:30,268 - stpipe - INFO - Results used jwst version: 1.16.1
2024-12-10 20:24:30,322 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:30,332 - stpipe - INFO - PARS-IMAGE2PIPELINE parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-image2pipeline_0002.asdf
2024-12-10 20:24:30,344 - stpipe.Image2Pipeline - INFO - Image2Pipeline instance created.
2024-12-10 20:24:30,345 - stpipe.Image2Pipeline.bkg_subtract - INFO - BackgroundStep instance created.
2024-12-10 20:24:30,346 - stpipe.Image2Pipeline.assign_wcs - INFO - AssignWcsStep instance created.
2024-12-10 20:24:30,347 - stpipe.Image2Pipeline.flat_field - INFO - FlatFieldStep instance created.
2024-12-10 20:24:30,348 - stpipe.Image2Pipeline.photom - INFO - PhotomStep instance created.
2024-12-10 20:24:30,349 - stpipe.Image2Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:30,550 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline running with args ('detector1/jw01475006001_02201_00017_nis_rate.fits',).
2024-12-10 20:24:30,558 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image2/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_bsub: False
steps:
bkg_subtract:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
save_combined_background: False
sigma: 3.0
maxiters: None
wfss_mmag_extract: None
assign_wcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
slit_y_low: -0.55
slit_y_high: 0.55
flat_field:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_interpolated_flat: False
user_supplied_flat: None
inverse: False
photom:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
inverse: False
source_type: None
mrs_time_correction: True
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: True
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
2024-12-10 20:24:30,613 - stpipe.Image2Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00017_nis_rate.fits' reftypes = ['area', 'camera', 'collimator', 'dflat', 'disperser', 'distortion', 'fflat', 'filteroffset', 'flat', 'fore', 'fpa', 'ifufore', 'ifupost', 'ifuslicer', 'msa', 'ote', 'photom', 'regions', 'sflat', 'specwcs', 'wavelengthrange']
2024-12-10 20:24:30,616 - stpipe.Image2Pipeline - INFO - Prefetch for AREA reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits'.
2024-12-10 20:24:30,617 - stpipe.Image2Pipeline - INFO - Prefetch for CAMERA reference file is 'N/A'.
2024-12-10 20:24:30,617 - stpipe.Image2Pipeline - INFO - Prefetch for COLLIMATOR reference file is 'N/A'.
2024-12-10 20:24:30,618 - stpipe.Image2Pipeline - INFO - Prefetch for DFLAT reference file is 'N/A'.
2024-12-10 20:24:30,618 - stpipe.Image2Pipeline - INFO - Prefetch for DISPERSER reference file is 'N/A'.
2024-12-10 20:24:30,619 - stpipe.Image2Pipeline - INFO - Prefetch for DISTORTION reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_distortion_0047.asdf'.
2024-12-10 20:24:30,619 - stpipe.Image2Pipeline - INFO - Prefetch for FFLAT reference file is 'N/A'.
2024-12-10 20:24:30,619 - stpipe.Image2Pipeline - INFO - Prefetch for FILTEROFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_filteroffset_0006.asdf'.
2024-12-10 20:24:30,620 - stpipe.Image2Pipeline - INFO - Prefetch for FLAT reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits'.
2024-12-10 20:24:30,621 - stpipe.Image2Pipeline - INFO - Prefetch for FORE reference file is 'N/A'.
2024-12-10 20:24:30,622 - stpipe.Image2Pipeline - INFO - Prefetch for FPA reference file is 'N/A'.
2024-12-10 20:24:30,622 - stpipe.Image2Pipeline - INFO - Prefetch for IFUFORE reference file is 'N/A'.
2024-12-10 20:24:30,623 - stpipe.Image2Pipeline - INFO - Prefetch for IFUPOST reference file is 'N/A'.
2024-12-10 20:24:30,623 - stpipe.Image2Pipeline - INFO - Prefetch for IFUSLICER reference file is 'N/A'.
2024-12-10 20:24:30,623 - stpipe.Image2Pipeline - INFO - Prefetch for MSA reference file is 'N/A'.
2024-12-10 20:24:30,624 - stpipe.Image2Pipeline - INFO - Prefetch for OTE reference file is 'N/A'.
2024-12-10 20:24:30,624 - stpipe.Image2Pipeline - INFO - Prefetch for PHOTOM reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits'.
2024-12-10 20:24:30,625 - stpipe.Image2Pipeline - INFO - Prefetch for REGIONS reference file is 'N/A'.
2024-12-10 20:24:30,626 - stpipe.Image2Pipeline - INFO - Prefetch for SFLAT reference file is 'N/A'.
2024-12-10 20:24:30,626 - stpipe.Image2Pipeline - INFO - Prefetch for SPECWCS reference file is 'N/A'.
2024-12-10 20:24:30,627 - stpipe.Image2Pipeline - INFO - Prefetch for WAVELENGTHRANGE reference file is 'N/A'.
2024-12-10 20:24:30,628 - stpipe.Image2Pipeline - INFO - Starting calwebb_image2 ...
2024-12-10 20:24:30,628 - stpipe.Image2Pipeline - INFO - Processing product detector1/jw01475006001_02201_00017_nis
2024-12-10 20:24:30,629 - stpipe.Image2Pipeline - INFO - Working on input detector1/jw01475006001_02201_00017_nis_rate.fits ...
2024-12-10 20:24:30,879 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00017_nis_image2pipeline.fits>,).
2024-12-10 20:24:31,061 - stpipe.Image2Pipeline.assign_wcs - INFO - Offsets from filteroffset file are 0.0, 0.0
2024-12-10 20:24:31,121 - stpipe.Image2Pipeline.assign_wcs - INFO - Update S_REGION to POLYGON ICRS 303.744319371 -26.791902974 303.784967553 -26.782816011 303.774662848 -26.746838158 303.734052767 -26.755906819
2024-12-10 20:24:31,121 - stpipe.Image2Pipeline.assign_wcs - INFO - assign_wcs updated S_REGION to POLYGON ICRS 303.744319371 -26.791902974 303.784967553 -26.782816011 303.774662848 -26.746838158 303.734052767 -26.755906819
2024-12-10 20:24:31,122 - stpipe.Image2Pipeline.assign_wcs - INFO - COMPLETED assign_wcs
2024-12-10 20:24:31,124 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/astropy/units/format/__init__.py:48: AstropyDeprecationWarning: The class "Fits" has been renamed to "FITS" in version 7.0. The old name is deprecated and may be removed in a future version.
Use FITS instead.
warnings.warn(
2024-12-10 20:24:31,125 - stpipe.Image2Pipeline.assign_wcs - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/gwcs/wcs.py:2035: AstropyDeprecationWarning: The get_format_name function is deprecated and may be removed in a future version.
Use to_string() instead.
cunit = frame.unit[fidx].get_format_name(u.format.Fits).upper()
2024-12-10 20:24:31,177 - stpipe.Image2Pipeline.assign_wcs - INFO - Step assign_wcs done
2024-12-10 20:24:31,373 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00017_nis_image2pipeline.fits>,).
2024-12-10 20:24:31,458 - stpipe.Image2Pipeline.flat_field - INFO - Using FLAT reference file: crds_cache/references/jwst/niriss/jwst_niriss_flat_0282.fits
2024-12-10 20:24:31,459 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type FFLAT
2024-12-10 20:24:31,460 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type SFLAT
2024-12-10 20:24:31,460 - stpipe.Image2Pipeline.flat_field - INFO - No reference found for type DFLAT
2024-12-10 20:24:31,619 - stpipe.Image2Pipeline.flat_field - INFO - Step flat_field done
2024-12-10 20:24:31,818 - stpipe.Image2Pipeline.photom - INFO - Step photom running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00017_nis_image2pipeline.fits>,).
2024-12-10 20:24:31,842 - stpipe.Image2Pipeline.photom - INFO - Using photom reference file: crds_cache/references/jwst/niriss/jwst_niriss_photom_0043.fits
2024-12-10 20:24:31,843 - stpipe.Image2Pipeline.photom - INFO - Using area reference file: crds_cache/references/jwst/niriss/jwst_niriss_area_0021.fits
2024-12-10 20:24:31,899 - stpipe.Image2Pipeline.photom - INFO - Using instrument: NIRISS
2024-12-10 20:24:31,899 - stpipe.Image2Pipeline.photom - INFO - detector: NIS
2024-12-10 20:24:31,900 - stpipe.Image2Pipeline.photom - INFO - exp_type: NIS_IMAGE
2024-12-10 20:24:31,901 - stpipe.Image2Pipeline.photom - INFO - filter: CLEAR
2024-12-10 20:24:31,901 - stpipe.Image2Pipeline.photom - INFO - pupil: F150W
2024-12-10 20:24:31,928 - stpipe.Image2Pipeline.photom - INFO - Pixel area map copied to output.
2024-12-10 20:24:31,929 - stpipe.Image2Pipeline.photom - INFO - Values for PIXAR_SR and PIXAR_A2 obtained from AREA reference file.
2024-12-10 20:24:31,930 - stpipe.Image2Pipeline.photom - INFO - PHOTMJSR value: 0.256732
2024-12-10 20:24:31,971 - stpipe.Image2Pipeline.photom - INFO - Step photom done
2024-12-10 20:24:32,171 - stpipe.Image2Pipeline.resample - INFO - Step resample running with args (<ImageModel(2048, 2048) from image2/jw01475006001_02201_00017_nis_image2pipeline.fits>,).
2024-12-10 20:24:32,172 - stpipe.Image2Pipeline.resample - INFO - Step skipped.
2024-12-10 20:24:32,174 - stpipe.Image2Pipeline - INFO - Finished processing product detector1/jw01475006001_02201_00017_nis
2024-12-10 20:24:32,174 - stpipe.Image2Pipeline - INFO - ... ending calwebb_image2
2024-12-10 20:24:32,175 - stpipe.Image2Pipeline - INFO - Results used CRDS context: jwst_1303.pmap
2024-12-10 20:24:32,443 - stpipe.Image2Pipeline - INFO - Saved model in image2/jw01475006001_02201_00017_nis_cal.fits
2024-12-10 20:24:32,444 - stpipe.Image2Pipeline - INFO - Step Image2Pipeline done
2024-12-10 20:24:32,445 - stpipe - INFO - Results used jwst version: 1.16.1
Identify *_cal.fits files#
cal_files = sorted(glob.glob(os.path.join(image2_dir, '*_cal.fits')))
Verify which pipeline steps were run#
cal_f = datamodels.open(cal_files[0])
cal_f.meta.cal_step.instance
{'assign_wcs': 'COMPLETE',
'charge_migration': 'COMPLETE',
'clean_flicker_noise': 'SKIPPED',
'dark_sub': 'COMPLETE',
'dq_init': 'COMPLETE',
'flat_field': 'COMPLETE',
'gain_scale': 'SKIPPED',
'group_scale': 'SKIPPED',
'ipc': 'SKIPPED',
'jump': 'COMPLETE',
'linearity': 'COMPLETE',
'persistence': 'SKIPPED',
'photom': 'COMPLETE',
'ramp_fit': 'COMPLETE',
'refpix': 'COMPLETE',
'resample': 'SKIPPED',
'saturation': 'COMPLETE',
'superbias': 'COMPLETE'}
Check which reference files were used to calibrate the dataset:
cal_f.meta.ref_file.instance
{'area': {'name': 'crds://jwst_niriss_area_0021.fits'},
'camera': {'name': 'N/A'},
'collimator': {'name': 'N/A'},
'crds': {'context_used': 'jwst_1303.pmap', 'sw_version': '12.0.8'},
'dark': {'name': 'crds://jwst_niriss_dark_0169.fits'},
'dflat': {'name': 'N/A'},
'disperser': {'name': 'N/A'},
'distortion': {'name': 'crds://jwst_niriss_distortion_0047.asdf'},
'fflat': {'name': 'N/A'},
'filteroffset': {'name': 'crds://jwst_niriss_filteroffset_0006.asdf'},
'flat': {'name': 'crds://jwst_niriss_flat_0282.fits'},
'fore': {'name': 'N/A'},
'fpa': {'name': 'N/A'},
'gain': {'name': 'crds://jwst_niriss_gain_0006.fits'},
'ifufore': {'name': 'N/A'},
'ifupost': {'name': 'N/A'},
'ifuslicer': {'name': 'N/A'},
'linearity': {'name': 'crds://jwst_niriss_linearity_0017.fits'},
'mask': {'name': 'crds://jwst_niriss_mask_0017.fits'},
'msa': {'name': 'N/A'},
'ote': {'name': 'N/A'},
'photom': {'name': 'crds://jwst_niriss_photom_0043.fits'},
'readnoise': {'name': 'crds://jwst_niriss_readnoise_0005.fits'},
'regions': {'name': 'N/A'},
'saturation': {'name': 'crds://jwst_niriss_saturation_0015.fits'},
'sflat': {'name': 'N/A'},
'specwcs': {'name': 'N/A'},
'superbias': {'name': 'crds://jwst_niriss_superbias_0183.fits'},
'wavelengthrange': {'name': 'N/A'}}
Display cal image#
Visualize a calibrated image, using the dataset from the first dither position as an example.
# Create an Imviz instance and set up default viewer
imviz_cal = Imviz()
viewer_cal = imviz_cal.default_viewer
# Read in the science array for our visualization dataset:
cal_science = cal_f.data
# Load the dataset into Imviz
imviz_cal.load_data(cal_science)
# Visualize the dataset:
imviz_cal.show()

plotopt = imviz_cal.plugins['Plot Options']
plotopt.stretch_function = 'sqrt'
plotopt.image_colormap = 'Viridis'
plotopt.stretch_preset = '95%'
plotopt.zoom_radius = 1024
The viewer looks like this:
viewer_cal.save('./cal_science.png')
Image('./cal_science.png')
3. Stage 3 Processing #
In the Image3 stage of the pipeline, the individual *_cal.fits files for each of the dither positions are combined to one single distortion corrected image. First, an Association needs to be created to inform the pipeline that these individual exposures are linked together.
By default, the Image3 stage of the pipeline performs the following steps on NIRISS data:
tweakreg - creates source catalogs of pointlike sources for each input image. The source catalog for each input image is compared to each other to derive coordinate transforms to align the images relative to each other.
As of CRDS context
jwst_1156.pmapand later, thepars-tweakregparameter reference file for NIRISS performs an absolute astrometric correction to GAIA data release 3 by default (i.e., theabs_refcatparameter is set toGAIADR3). Though this default correction generally improves results compared with not doing this alignment, it can sometimes result in poor performance in crowded or sparse fields, so users are encouraged to check astrometric accuracy and revisit this step if necessary.As of pipeline version 1.12.5, the default source finding algorithm is
DAOStarFinderwhich can result in up to 0.5 pix uncertainties in the centroids for undersampled PSFs, like the NIRISS PSFs at short wavelengths (Goudfrooij 2022). There are plans to update the default algorithm toIRAFStarFinderin future pipeline versions.
skymatch - measures the background level from the sky to use as input into the subsequent
outlier detectionandresamplesteps.outlier detection - flags any remaining cosmic rays, bad pixels, or other artifacts not already flagged during the
DETECTOR1stage of the pipeline, using all input images to create a median image so that outliers in individual images can be identified.resample - resamples each input image based on its WCS and distortion information and creates a single undistorted image.
source catalog - creates a catalog of detected sources along with measured photometries and morphologies (i.e., point-like vs extended). Useful for quicklooks, but optimization is likely needed for specific science cases, which is an on-going investigation for the NIRISS team. Users may wish to experiment with changing the
snr_thresholdanddeblendoptions. Modifications to the following parameters will not significantly improve data quality and it is advised to keep them at their default values:aperture_ee1,aperture_ee2,aperture_ee3,ci1_star_threshold,ci2_star_threshold.
Create Association File#
An association file lists the exposures to calibrated together in Stage 3 of the pipeline. Note that an association file is available for download from MAST, with a filename of *_asn.json. Here we show how to create an association file to point to the data products created when processing data through the pipeline. Note that the output products will have a rootname that is specified by the product_name in the association file. For this tutorial, the rootname of the output products will be image3_association.
# Create a Level 3 Association
associations = asn_from_list.asn_from_list(cal_files,
rule=DMS_Level3_Base,
product_name='image3_association')
associations.data['asn_type'] = 'image3'
associations.data['program'] = pid
# Format association as .json file
asn_filename, serialized = associations.dump(format="json")
# Write out association file
with open(asn_filename, "w") as fd:
fd.write(serialized)
2024-12-10 20:24:33,409 - stpipe - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst/associations/association.py:215: UserWarning: Input association file contains path information; note that this can complicate usage and/or sharing of such files.
warnings.warn(err_str, UserWarning)
Run Image3 stage of the pipeline#
Given the grouped exposures in the association file, the Image3 stage of the pipeline will produce:
a
*_cr.fitsfile produced by theoutlier_detectionstep, where theDQarray marks the pixels flagged as outliers.a final combined, rectified image with name
*_i2d.fits,a source catalog with name
*_cat.ecsv,a segmentation map file (
*_segm.fits) which has integer values at the pixel locations where a source is detected where the pixel values match the source ID number in the catalog.
# Define directory to save output from Image2
image3_dir = 'image3/'
# Create directory if it does not exist
if not os.path.isdir(image3_dir):
os.mkdir(image3_dir)
# Run Stage 3
i2d_result = Image3Pipeline.call(asn_filename,
output_dir=image3_dir,
save_results=True)
2024-12-10 20:24:33,577 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-tweakregstep_0079.asdf 1.7 K bytes (1 / 1 files) (0 / 1.7 K bytes)
2024-12-10 20:24:33,687 - stpipe - INFO - PARS-TWEAKREGSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-tweakregstep_0079.asdf
2024-12-10 20:24:33,699 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-outlierdetectionstep_0010.asdf 5.3 K bytes (1 / 1 files) (0 / 5.3 K bytes)
2024-12-10 20:24:33,838 - stpipe - INFO - PARS-OUTLIERDETECTIONSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-outlierdetectionstep_0010.asdf
2024-12-10 20:24:33,853 - stpipe - INFO - PARS-RESAMPLESTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-resamplestep_0001.asdf
2024-12-10 20:24:33,862 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_pars-sourcecatalogstep_0011.asdf 1.3 K bytes (1 / 1 files) (0 / 1.3 K bytes)
2024-12-10 20:24:33,980 - stpipe - INFO - PARS-SOURCECATALOGSTEP parameters found: crds_cache/references/jwst/niriss/jwst_niriss_pars-sourcecatalogstep_0011.asdf
2024-12-10 20:24:33,996 - stpipe.Image3Pipeline - INFO - Image3Pipeline instance created.
2024-12-10 20:24:33,997 - stpipe.Image3Pipeline.assign_mtwcs - INFO - AssignMTWcsStep instance created.
2024-12-10 20:24:33,999 - stpipe.Image3Pipeline.tweakreg - INFO - TweakRegStep instance created.
2024-12-10 20:24:34,001 - stpipe.Image3Pipeline.skymatch - INFO - SkyMatchStep instance created.
2024-12-10 20:24:34,002 - stpipe.Image3Pipeline.outlier_detection - INFO - OutlierDetectionStep instance created.
2024-12-10 20:24:34,003 - stpipe.Image3Pipeline.resample - INFO - ResampleStep instance created.
2024-12-10 20:24:34,004 - stpipe.Image3Pipeline.source_catalog - INFO - SourceCatalogStep instance created.
2024-12-10 20:24:34,220 - stpipe.Image3Pipeline - INFO - Step Image3Pipeline running with args ('jw01475-a3001_image3_00019_asn.json',).
2024-12-10 20:24:34,234 - stpipe.Image3Pipeline - INFO - Step Image3Pipeline parameters are:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: image3/
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: True
skip: False
suffix: None
search_output_file: True
input_dir: ''
in_memory: True
steps:
assign_mtwcs:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: True
output_use_index: True
save_results: False
skip: False
suffix: assign_mtwcs
search_output_file: True
input_dir: ''
tweakreg:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: True
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
save_catalogs: False
use_custom_catalogs: False
catalog_format: ecsv
catfile: ''
starfinder: iraf
snr_threshold: 10
bkg_boxsize: 400
kernel_fwhm: 2.0
minsep_fwhm: 0.0
sigma_radius: 1.5
sharplo: 0.2
sharphi: 3.0
roundlo: -1.0
roundhi: 1.0
brightest: 100
peakmax: None
npixels: 10
connectivity: '8'
nlevels: 32
contrast: 0.001
multithresh_mode: exponential
localbkg_width: 0
apermask_method: correct
kron_params: None
enforce_user_order: False
expand_refcat: False
minobj: 10
fitgeometry: rshift
nclip: 3
sigma: 3.0
searchrad: 1.0
use2dhist: True
separation: 1.5
tolerance: 1.0
xoffset: 0.0
yoffset: 0.0
abs_refcat: GAIADR3
save_abs_catalog: False
abs_minobj: 15
abs_fitgeometry: rshift
abs_nclip: 3
abs_sigma: 3.0
abs_searchrad: 6.0
abs_use2dhist: True
abs_separation: 1.0
abs_tolerance: 0.7
sip_approx: True
sip_max_pix_error: 0.01
sip_degree: None
sip_max_inv_pix_error: 0.01
sip_inv_degree: None
sip_npoints: 12
in_memory: True
skymatch:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
skymethod: match
match_down: True
subtract: False
stepsize: None
skystat: mode
dqbits: ~DO_NOT_USE+NON_SCIENCE
lower: None
upper: None
nclip: 5
lsigma: 4.0
usigma: 4.0
binwidth: 0.1
in_memory: True
outlier_detection:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: False
input_dir: ''
weight_type: ivm
pixfrac: 1.0
kernel: square
fillval: INDEF
nlow: 0
nhigh: 0
maskpt: 0.7
snr: 5.0 4.0
scale: 2.1 0.7
backg: 0.0
kernel_size: 7 7
threshold_percent: 99.8
rolling_window_width: 25
ifu_second_check: False
save_intermediate_results: False
resample_data: True
good_bits: ~DO_NOT_USE
allowed_memory: None
in_memory: False
resample:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: None
search_output_file: True
input_dir: ''
pixfrac: 1.0
kernel: square
fillval: NAN
weight_type: ivm
output_shape: None
crpix: None
crval: None
rotation: None
pixel_scale_ratio: 1.0
pixel_scale: None
output_wcs: ''
single: False
blendheaders: True
allowed_memory: None
in_memory: True
source_catalog:
pre_hooks: []
post_hooks: []
output_file: None
output_dir: None
output_ext: .fits
output_use_model: False
output_use_index: True
save_results: False
skip: False
suffix: cat
search_output_file: True
input_dir: ''
bkg_boxsize: 100
kernel_fwhm: 3.0
snr_threshold: 3.0
npixels: 5
deblend: False
aperture_ee1: 50
aperture_ee2: 70
aperture_ee3: 80
ci1_star_threshold: 1.4
ci2_star_threshold: 1.4
2024-12-10 20:24:34,246 - stpipe.Image3Pipeline - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst/associations/association.py:215: UserWarning: Input association file contains path information; note that this can complicate usage and/or sharing of such files.
warnings.warn(err_str, UserWarning)
2024-12-10 20:24:34,382 - stpipe.Image3Pipeline - INFO - Prefetching reference files for dataset: 'jw01475006001_02201_00001_nis_cal.fits' reftypes = ['abvegaoffset', 'apcorr']
2024-12-10 20:24:34,385 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_abvegaoffset_0003.asdf 2.1 K bytes (1 / 2 files) (0 / 16.5 K bytes)
2024-12-10 20:24:34,497 - CRDS - INFO - Fetching crds_cache/references/jwst/niriss/jwst_niriss_apcorr_0008.fits 14.4 K bytes (2 / 2 files) (2.1 K / 16.5 K bytes)
2024-12-10 20:24:34,610 - stpipe.Image3Pipeline - INFO - Prefetch for ABVEGAOFFSET reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_abvegaoffset_0003.asdf'.
2024-12-10 20:24:34,611 - stpipe.Image3Pipeline - INFO - Prefetch for APCORR reference file is 'crds_cache/references/jwst/niriss/jwst_niriss_apcorr_0008.fits'.
2024-12-10 20:24:34,613 - stpipe.Image3Pipeline - INFO - Starting calwebb_image3 ...
2024-12-10 20:24:34,622 - stpipe.Image3Pipeline - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst/associations/association.py:215: UserWarning: Input association file contains path information; note that this can complicate usage and/or sharing of such files.
warnings.warn(err_str, UserWarning)
2024-12-10 20:24:35,194 - stpipe.Image3Pipeline.tweakreg - INFO - Step tweakreg running with args (<jwst.datamodels.library.ModelLibrary object at 0x7f1ec88e4290>,).
2024-12-10 20:24:36,589 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00001_nis_cal.fits.
2024-12-10 20:24:38,157 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00002_nis_cal.fits.
2024-12-10 20:24:39,733 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00003_nis_cal.fits.
2024-12-10 20:24:41,306 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00004_nis_cal.fits.
2024-12-10 20:24:42,893 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00005_nis_cal.fits.
2024-12-10 20:24:44,470 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00006_nis_cal.fits.
2024-12-10 20:24:46,033 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00007_nis_cal.fits.
2024-12-10 20:24:47,603 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00008_nis_cal.fits.
2024-12-10 20:24:49,180 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00009_nis_cal.fits.
2024-12-10 20:24:50,749 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00010_nis_cal.fits.
2024-12-10 20:24:52,568 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00011_nis_cal.fits.
2024-12-10 20:24:54,146 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00012_nis_cal.fits.
2024-12-10 20:24:55,771 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00013_nis_cal.fits.
2024-12-10 20:24:57,376 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00014_nis_cal.fits.
2024-12-10 20:24:58,995 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00015_nis_cal.fits.
2024-12-10 20:25:00,625 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00016_nis_cal.fits.
2024-12-10 20:25:02,266 - stpipe.Image3Pipeline.tweakreg - INFO - Detected 100 sources in jw01475006001_02201_00017_nis_cal.fits.
2024-12-10 20:25:02,293 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:02,294 - stpipe.Image3Pipeline.tweakreg - INFO - Number of image groups to be aligned: 17.
2024-12-10 20:25:02,294 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:02,295 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() started on 2024-12-10 20:25:02.294881
2024-12-10 20:25:02,296 - stpipe.Image3Pipeline.tweakreg - INFO - Version 0.8.9
2024-12-10 20:25:02,296 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:22,464 - stpipe.Image3Pipeline.tweakreg - INFO - Selected image 'GROUP ID: jw01475006001_02201_17' as reference image
2024-12-10 20:25:22,471 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_12' to the reference catalog.
2024-12-10 20:25:22,521 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00012_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00017_nis_cal' catalog.
2024-12-10 20:25:22,522 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:22,523 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.05024, 0.049 (arcsec) with significance of 48.83 and 53 matches.
2024-12-10 20:25:22,524 - stpipe.Image3Pipeline.tweakreg - INFO - Found 52 matches for 'GROUP ID: jw01475006001_02201_12'...
2024-12-10 20:25:22,525 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:22,528 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_12:
2024-12-10 20:25:22,528 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00297273 YSH: -0.00187108 ROT: -0.000591114 SCALE: 1
2024-12-10 20:25:22,529 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:22,529 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.00789271 FIT MAE: 0.00615887
2024-12-10 20:25:22,530 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 50 objects.
2024-12-10 20:25:22,579 - stpipe.Image3Pipeline.tweakreg - INFO - Added 48 unmatched sources from 'GROUP ID: jw01475006001_02201_12' to the reference catalog.
2024-12-10 20:25:24,628 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_8' to the reference catalog.
2024-12-10 20:25:24,676 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00008_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00012_nis_cal' catalog.
2024-12-10 20:25:24,677 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:24,678 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.049, 0.049 (arcsec) with significance of 43.82 and 50 matches.
2024-12-10 20:25:24,680 - stpipe.Image3Pipeline.tweakreg - INFO - Found 48 matches for 'GROUP ID: jw01475006001_02201_8'...
2024-12-10 20:25:24,680 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:24,683 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_8:
2024-12-10 20:25:24,683 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00164925 YSH: -0.00369618 ROT: 0.0011737 SCALE: 1
2024-12-10 20:25:24,684 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:24,684 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.01276 FIT MAE: 0.0076927
2024-12-10 20:25:24,685 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 45 objects.
2024-12-10 20:25:24,729 - stpipe.Image3Pipeline.tweakreg - INFO - Added 52 unmatched sources from 'GROUP ID: jw01475006001_02201_8' to the reference catalog.
2024-12-10 20:25:26,592 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_13' to the reference catalog.
2024-12-10 20:25:26,646 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00013_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00008_nis_cal' catalog.
2024-12-10 20:25:26,647 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:26,649 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.04796, 0.04692 (arcsec) with significance of 51.17 and 63 matches.
2024-12-10 20:25:26,650 - stpipe.Image3Pipeline.tweakreg - INFO - Found 58 matches for 'GROUP ID: jw01475006001_02201_13'...
2024-12-10 20:25:26,651 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:26,654 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_13:
2024-12-10 20:25:26,654 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00303404 YSH: -0.00194698 ROT: 7.34316e-05 SCALE: 1
2024-12-10 20:25:26,655 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:26,656 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.00929328 FIT MAE: 0.00667297
2024-12-10 20:25:26,656 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 56 objects.
2024-12-10 20:25:26,701 - stpipe.Image3Pipeline.tweakreg - INFO - Added 42 unmatched sources from 'GROUP ID: jw01475006001_02201_13' to the reference catalog.
2024-12-10 20:25:28,766 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_14' to the reference catalog.
2024-12-10 20:25:28,819 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00014_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00013_nis_cal' catalog.
2024-12-10 20:25:28,819 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:28,821 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.04707, 0.05382 (arcsec) with significance of 52.42 and 68 matches.
2024-12-10 20:25:28,822 - stpipe.Image3Pipeline.tweakreg - INFO - Found 56 matches for 'GROUP ID: jw01475006001_02201_14'...
2024-12-10 20:25:28,823 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:28,826 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_14:
2024-12-10 20:25:28,826 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.0122021 YSH: 0.00041192 ROT: -0.00193315 SCALE: 1
2024-12-10 20:25:28,827 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:28,827 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0626783 FIT MAE: 0.0179162
2024-12-10 20:25:28,827 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 54 objects.
2024-12-10 20:25:28,873 - stpipe.Image3Pipeline.tweakreg - INFO - Added 44 unmatched sources from 'GROUP ID: jw01475006001_02201_14' to the reference catalog.
2024-12-10 20:25:30,704 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_16' to the reference catalog.
2024-12-10 20:25:30,757 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00016_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00014_nis_cal' catalog.
2024-12-10 20:25:30,758 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:30,760 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.05087, 0.04619 (arcsec) with significance of 55.78 and 70 matches.
2024-12-10 20:25:30,761 - stpipe.Image3Pipeline.tweakreg - INFO - Found 62 matches for 'GROUP ID: jw01475006001_02201_16'...
2024-12-10 20:25:30,762 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:30,764 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_16:
2024-12-10 20:25:30,765 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00592855 YSH: -0.000195081 ROT: -0.000676598 SCALE: 1
2024-12-10 20:25:30,765 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:30,766 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0157021 FIT MAE: 0.00707357
2024-12-10 20:25:30,766 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 59 objects.
2024-12-10 20:25:31,097 - stpipe.Image3Pipeline.tweakreg - INFO - Added 38 unmatched sources from 'GROUP ID: jw01475006001_02201_16' to the reference catalog.
2024-12-10 20:25:32,953 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_11' to the reference catalog.
2024-12-10 20:25:33,005 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00011_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00016_nis_cal' catalog.
2024-12-10 20:25:33,005 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:33,007 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.049, 0.049 (arcsec) with significance of 58.13 and 70 matches.
2024-12-10 20:25:33,008 - stpipe.Image3Pipeline.tweakreg - INFO - Found 63 matches for 'GROUP ID: jw01475006001_02201_11'...
2024-12-10 20:25:33,009 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:33,012 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_11:
2024-12-10 20:25:33,012 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00958002 YSH: 0.00354008 ROT: 0.0104241 SCALE: 1
2024-12-10 20:25:33,013 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:33,013 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.062213 FIT MAE: 0.0205152
2024-12-10 20:25:33,014 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 60 objects.
2024-12-10 20:25:33,061 - stpipe.Image3Pipeline.tweakreg - INFO - Added 37 unmatched sources from 'GROUP ID: jw01475006001_02201_11' to the reference catalog.
2024-12-10 20:25:34,963 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_10' to the reference catalog.
2024-12-10 20:25:35,018 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00010_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00011_nis_cal' catalog.
2024-12-10 20:25:35,019 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:35,021 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.049, 0.05155 (arcsec) with significance of 49.16 and 77 matches.
2024-12-10 20:25:35,022 - stpipe.Image3Pipeline.tweakreg - INFO - Found 67 matches for 'GROUP ID: jw01475006001_02201_10'...
2024-12-10 20:25:35,022 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:35,025 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_10:
2024-12-10 20:25:35,026 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00464647 YSH: 0.000554522 ROT: 0.000197908 SCALE: 1
2024-12-10 20:25:35,026 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:35,027 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0107048 FIT MAE: 0.0082514
2024-12-10 20:25:35,027 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 66 objects.
2024-12-10 20:25:35,075 - stpipe.Image3Pipeline.tweakreg - INFO - Added 33 unmatched sources from 'GROUP ID: jw01475006001_02201_10' to the reference catalog.
2024-12-10 20:25:36,747 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_15' to the reference catalog.
2024-12-10 20:25:36,800 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00015_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00010_nis_cal' catalog.
2024-12-10 20:25:36,800 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:36,803 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of -0.02334, -0.03547 (arcsec) with significance of 55.49 and 80 matches.
2024-12-10 20:25:36,804 - stpipe.Image3Pipeline.tweakreg - INFO - Found 65 matches for 'GROUP ID: jw01475006001_02201_15'...
2024-12-10 20:25:36,805 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:36,807 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_15:
2024-12-10 20:25:36,808 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00598402 YSH: 0.00242701 ROT: -0.00091101 SCALE: 1
2024-12-10 20:25:36,808 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:36,809 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0575169 FIT MAE: 0.0172465
2024-12-10 20:25:36,810 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 62 objects.
2024-12-10 20:25:36,859 - stpipe.Image3Pipeline.tweakreg - INFO - Added 35 unmatched sources from 'GROUP ID: jw01475006001_02201_15' to the reference catalog.
2024-12-10 20:25:38,352 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_1' to the reference catalog.
2024-12-10 20:25:38,404 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00001_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00015_nis_cal' catalog.
2024-12-10 20:25:38,405 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:38,406 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.04976, 0.04595 (arcsec) with significance of 57.75 and 86 matches.
2024-12-10 20:25:38,407 - stpipe.Image3Pipeline.tweakreg - INFO - Found 65 matches for 'GROUP ID: jw01475006001_02201_1'...
2024-12-10 20:25:38,408 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:38,410 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_1:
2024-12-10 20:25:38,411 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00215817 YSH: 0.000376617 ROT: -0.000540888 SCALE: 1
2024-12-10 20:25:38,411 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:38,412 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.00783033 FIT MAE: 0.00654007
2024-12-10 20:25:38,412 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 63 objects.
2024-12-10 20:25:38,459 - stpipe.Image3Pipeline.tweakreg - INFO - Added 35 unmatched sources from 'GROUP ID: jw01475006001_02201_1' to the reference catalog.
2024-12-10 20:25:39,792 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_9' to the reference catalog.
2024-12-10 20:25:39,843 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00009_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00001_nis_cal' catalog.
2024-12-10 20:25:39,843 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:39,845 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.04984, 0.04816 (arcsec) with significance of 50.91 and 78 matches.
2024-12-10 20:25:39,846 - stpipe.Image3Pipeline.tweakreg - INFO - Found 59 matches for 'GROUP ID: jw01475006001_02201_9'...
2024-12-10 20:25:39,847 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:39,849 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_9:
2024-12-10 20:25:39,849 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00529789 YSH: -0.0018275 ROT: -0.00100167 SCALE: 1
2024-12-10 20:25:39,850 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:39,851 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.00772268 FIT MAE: 0.00650076
2024-12-10 20:25:39,851 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 57 objects.
2024-12-10 20:25:39,898 - stpipe.Image3Pipeline.tweakreg - INFO - Added 41 unmatched sources from 'GROUP ID: jw01475006001_02201_9' to the reference catalog.
2024-12-10 20:25:41,190 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_4' to the reference catalog.
2024-12-10 20:25:41,239 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00004_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00009_nis_cal' catalog.
2024-12-10 20:25:41,240 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:41,242 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.049, 0.04975 (arcsec) with significance of 59.63 and 87 matches.
2024-12-10 20:25:41,244 - stpipe.Image3Pipeline.tweakreg - INFO - Found 62 matches for 'GROUP ID: jw01475006001_02201_4'...
2024-12-10 20:25:41,244 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:41,247 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_4:
2024-12-10 20:25:41,248 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00574765 YSH: 0.000846227 ROT: -0.0092664 SCALE: 1
2024-12-10 20:25:41,248 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:41,248 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0612653 FIT MAE: 0.020322
2024-12-10 20:25:41,249 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 60 objects.
2024-12-10 20:25:41,297 - stpipe.Image3Pipeline.tweakreg - INFO - Added 38 unmatched sources from 'GROUP ID: jw01475006001_02201_4' to the reference catalog.
2024-12-10 20:25:42,318 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_5' to the reference catalog.
2024-12-10 20:25:42,368 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00005_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00004_nis_cal' catalog.
2024-12-10 20:25:42,368 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:42,370 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.049, 0.049 (arcsec) with significance of 55.76 and 94 matches.
2024-12-10 20:25:42,372 - stpipe.Image3Pipeline.tweakreg - INFO - Found 59 matches for 'GROUP ID: jw01475006001_02201_5'...
2024-12-10 20:25:42,372 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:42,375 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_5:
2024-12-10 20:25:42,375 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.0125868 YSH: 0.00303293 ROT: -0.00240757 SCALE: 1
2024-12-10 20:25:42,376 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:42,376 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0651953 FIT MAE: 0.0191239
2024-12-10 20:25:42,377 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 56 objects.
2024-12-10 20:25:42,424 - stpipe.Image3Pipeline.tweakreg - INFO - Added 41 unmatched sources from 'GROUP ID: jw01475006001_02201_5' to the reference catalog.
2024-12-10 20:25:43,111 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_6' to the reference catalog.
2024-12-10 20:25:43,164 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00006_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00005_nis_cal' catalog.
2024-12-10 20:25:43,165 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:43,166 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.0477, 0.04965 (arcsec) with significance of 68.6 and 101 matches.
2024-12-10 20:25:43,168 - stpipe.Image3Pipeline.tweakreg - INFO - Found 64 matches for 'GROUP ID: jw01475006001_02201_6'...
2024-12-10 20:25:43,168 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:43,171 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_6:
2024-12-10 20:25:43,171 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -5.64577e-07 YSH: 0.00570035 ROT: 0.000500578 SCALE: 1
2024-12-10 20:25:43,171 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:43,173 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.00778831 FIT MAE: 0.00590413
2024-12-10 20:25:43,173 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 60 objects.
2024-12-10 20:25:43,220 - stpipe.Image3Pipeline.tweakreg - INFO - Added 36 unmatched sources from 'GROUP ID: jw01475006001_02201_6' to the reference catalog.
2024-12-10 20:25:43,727 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_2' to the reference catalog.
2024-12-10 20:25:43,783 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00002_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00006_nis_cal' catalog.
2024-12-10 20:25:43,784 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:43,785 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.04767, 0.03219 (arcsec) with significance of 58.68 and 103 matches.
2024-12-10 20:25:43,787 - stpipe.Image3Pipeline.tweakreg - INFO - Found 69 matches for 'GROUP ID: jw01475006001_02201_2'...
2024-12-10 20:25:43,788 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:43,790 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_2:
2024-12-10 20:25:43,791 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00427682 YSH: 0.00472328 ROT: 0.00121112 SCALE: 1
2024-12-10 20:25:43,791 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:43,792 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0588941 FIT MAE: 0.0159931
2024-12-10 20:25:43,792 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 66 objects.
2024-12-10 20:25:43,838 - stpipe.Image3Pipeline.tweakreg - INFO - Added 31 unmatched sources from 'GROUP ID: jw01475006001_02201_2' to the reference catalog.
2024-12-10 20:25:44,157 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_3' to the reference catalog.
2024-12-10 20:25:44,207 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00003_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00002_nis_cal' catalog.
2024-12-10 20:25:44,207 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:44,209 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.05121, 0.04605 (arcsec) with significance of 41.57 and 89 matches.
2024-12-10 20:25:44,210 - stpipe.Image3Pipeline.tweakreg - INFO - Found 52 matches for 'GROUP ID: jw01475006001_02201_3'...
2024-12-10 20:25:44,211 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:44,214 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_3:
2024-12-10 20:25:44,214 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: 0.00295282 YSH: -0.00595903 ROT: 0.000161523 SCALE: 1
2024-12-10 20:25:44,214 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:44,215 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0696149 FIT MAE: 0.0210842
2024-12-10 20:25:44,216 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 50 objects.
2024-12-10 20:25:44,261 - stpipe.Image3Pipeline.tweakreg - INFO - Added 48 unmatched sources from 'GROUP ID: jw01475006001_02201_3' to the reference catalog.
2024-12-10 20:25:44,401 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: jw01475006001_02201_7' to the reference catalog.
2024-12-10 20:25:44,453 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_00007_nis_cal' catalog with sources from the reference 'jw01475006001_02201_00003_nis_cal' catalog.
2024-12-10 20:25:44,454 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:25:44,456 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of -0.07161, 0.01078 (arcsec) with significance of 52.26 and 103 matches.
2024-12-10 20:25:44,457 - stpipe.Image3Pipeline.tweakreg - INFO - Found 65 matches for 'GROUP ID: jw01475006001_02201_7'...
2024-12-10 20:25:44,458 - stpipe.Image3Pipeline.tweakreg - INFO - Performing 'rshift' fit
2024-12-10 20:25:44,460 - stpipe.Image3Pipeline.tweakreg - INFO - Computed 'rshift' fit for GROUP ID: jw01475006001_02201_7:
2024-12-10 20:25:44,461 - stpipe.Image3Pipeline.tweakreg - INFO - XSH: -0.00894993 YSH: 0.00463131 ROT: -0.0028708 SCALE: 1
2024-12-10 20:25:44,461 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:44,462 - stpipe.Image3Pipeline.tweakreg - INFO - FIT RMSE: 0.0940692 FIT MAE: 0.023655
2024-12-10 20:25:44,462 - stpipe.Image3Pipeline.tweakreg - INFO - Final solution based on 60 objects.
2024-12-10 20:25:44,508 - stpipe.Image3Pipeline.tweakreg - INFO - Added 36 unmatched sources from 'GROUP ID: jw01475006001_02201_7' to the reference catalog.
2024-12-10 20:25:44,509 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:44,509 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() ended on 2024-12-10 20:25:44.509290
2024-12-10 20:25:44,510 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() TOTAL RUN TIME: 0:00:42.214409
2024-12-10 20:25:44,510 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:45,375 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:25:45,375 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() started on 2024-12-10 20:25:45.375210
2024-12-10 20:25:45,376 - stpipe.Image3Pipeline.tweakreg - INFO - Version 0.8.9
2024-12-10 20:25:45,377 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:27:36,327 - stpipe.Image3Pipeline.tweakreg - INFO - Aligning image catalog 'GROUP ID: 987654' to the reference catalog.
2024-12-10 20:27:36,385 - stpipe.Image3Pipeline.tweakreg - INFO - Matching sources from 'jw01475006001_02201_000' catalog with sources from the reference 'Unnamed' catalog.
2024-12-10 20:27:36,386 - stpipe.Image3Pipeline.tweakreg - INFO - Computing initial guess for X and Y shifts...
2024-12-10 20:27:36,389 - stpipe.Image3Pipeline.tweakreg - INFO - Found initial X and Y shifts of 0.1692, 0.002131 (arcsec) with significance of 10.44 and 31 matches.
2024-12-10 20:27:36,390 - stpipe.Image3Pipeline.tweakreg - INFO - Found 13 matches for 'GROUP ID: 987654'...
2024-12-10 20:27:36,391 - stpipe.Image3Pipeline.tweakreg - WARNING - Not enough matches (< 13) found for image catalog 'GROUP ID: 987654'.
2024-12-10 20:27:36,392 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:27:36,392 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() ended on 2024-12-10 20:27:36.392137
2024-12-10 20:27:36,393 - stpipe.Image3Pipeline.tweakreg - INFO - ***** tweakwcs.imalign.align_wcs() TOTAL RUN TIME: 0:01:51.016927
2024-12-10 20:27:36,393 - stpipe.Image3Pipeline.tweakreg - INFO -
2024-12-10 20:27:36,407 - stpipe.Image3Pipeline.tweakreg - INFO - Step tweakreg done
2024-12-10 20:27:36,760 - stpipe.Image3Pipeline.skymatch - INFO - Step skymatch running with args (<jwst.datamodels.library.ModelLibrary object at 0x7f1ec88e4290>,).
2024-12-10 20:27:37,096 - stpipe.Image3Pipeline.skymatch - INFO -
2024-12-10 20:27:37,097 - stpipe.Image3Pipeline.skymatch - INFO - ***** jwst.skymatch.skymatch.match() started on 2024-12-10 20:27:37.096575
2024-12-10 20:27:37,097 - stpipe.Image3Pipeline.skymatch - INFO -
2024-12-10 20:27:37,098 - stpipe.Image3Pipeline.skymatch - INFO - Sky computation method: 'match'
2024-12-10 20:27:37,098 - stpipe.Image3Pipeline.skymatch - INFO - Sky matching direction: DOWN
2024-12-10 20:27:37,098 - stpipe.Image3Pipeline.skymatch - INFO - Sky subtraction from image data: OFF
2024-12-10 20:27:37,099 - stpipe.Image3Pipeline.skymatch - INFO -
2024-12-10 20:27:37,099 - stpipe.Image3Pipeline.skymatch - INFO - ---- Computing differences in sky values in overlapping regions.
2024-12-10 20:29:53,714 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00001_nis_cal.fits. Sky background: 0.000777844
2024-12-10 20:29:53,715 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00002_nis_cal.fits. Sky background: 0
2024-12-10 20:29:53,715 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00003_nis_cal.fits. Sky background: 0.00194414
2024-12-10 20:29:53,716 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00004_nis_cal.fits. Sky background: 0.00261598
2024-12-10 20:29:53,716 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00005_nis_cal.fits. Sky background: 0.00184969
2024-12-10 20:29:53,717 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00006_nis_cal.fits. Sky background: 0.00106297
2024-12-10 20:29:53,717 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00007_nis_cal.fits. Sky background: 0.000463979
2024-12-10 20:29:53,718 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00008_nis_cal.fits. Sky background: 0.00158776
2024-12-10 20:29:53,718 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00009_nis_cal.fits. Sky background: 0.00102022
2024-12-10 20:29:53,718 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00010_nis_cal.fits. Sky background: 0.00180598
2024-12-10 20:29:53,719 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00011_nis_cal.fits. Sky background: 0.000597871
2024-12-10 20:29:53,719 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00012_nis_cal.fits. Sky background: 0.000848144
2024-12-10 20:29:53,720 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00013_nis_cal.fits. Sky background: 0.00153453
2024-12-10 20:29:53,720 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00014_nis_cal.fits. Sky background: 0.00252335
2024-12-10 20:29:53,720 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00015_nis_cal.fits. Sky background: 0.000937484
2024-12-10 20:29:53,721 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00016_nis_cal.fits. Sky background: 0.00180074
2024-12-10 20:29:53,721 - stpipe.Image3Pipeline.skymatch - INFO - * Image ID=jw01475006001_02201_00017_nis_cal.fits. Sky background: 0.00180182
2024-12-10 20:29:53,722 - stpipe.Image3Pipeline.skymatch - INFO -
2024-12-10 20:29:53,722 - stpipe.Image3Pipeline.skymatch - INFO - ***** jwst.skymatch.skymatch.match() ended on 2024-12-10 20:29:53.722045
2024-12-10 20:29:53,722 - stpipe.Image3Pipeline.skymatch - INFO - ***** jwst.skymatch.skymatch.match() TOTAL RUN TIME: 0:02:16.625470
2024-12-10 20:29:53,723 - stpipe.Image3Pipeline.skymatch - INFO -
2024-12-10 20:29:53,761 - stpipe.Image3Pipeline.skymatch - INFO - Step skymatch done
2024-12-10 20:29:54,057 - stpipe.Image3Pipeline.outlier_detection - INFO - Step outlier_detection running with args (<jwst.datamodels.library.ModelLibrary object at 0x7f1ec88e4290>,).
2024-12-10 20:29:54,058 - stpipe.Image3Pipeline.outlier_detection - INFO - Outlier Detection mode: imaging
2024-12-10 20:29:54,060 - stpipe.Image3Pipeline.outlier_detection - INFO - Outlier Detection asn_id: a3001
2024-12-10 20:29:54,060 - stpipe.Image3Pipeline.outlier_detection - INFO - Driz parameter kernel: square
2024-12-10 20:29:54,061 - stpipe.Image3Pipeline.outlier_detection - INFO - Driz parameter pixfrac: 1.0
2024-12-10 20:29:54,061 - stpipe.Image3Pipeline.outlier_detection - INFO - Driz parameter fillval: INDEF
2024-12-10 20:29:54,061 - stpipe.Image3Pipeline.outlier_detection - INFO - Driz parameter weight_type: ivm
2024-12-10 20:29:54,062 - stpipe.Image3Pipeline.outlier_detection - INFO - Output pixel scale ratio: 1.0
2024-12-10 20:29:54,209 - stpipe.Image3Pipeline.outlier_detection - INFO - Computed output pixel scale: 0.06556276592712629 arcsec.
2024-12-10 20:29:54,269 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:29:54,368 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:29:56,894 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:29:57,897 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00001_nis_a3001_outlier_i2d.fits
2024-12-10 20:29:57,906 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:29:57,973 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:00,097 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:01,156 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00002_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:01,165 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:01,229 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:03,328 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:04,322 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00003_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:04,331 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:04,395 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:06,476 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:07,478 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00004_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:07,486 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:07,550 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:09,614 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:10,606 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00005_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:10,614 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:10,678 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:12,785 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:13,782 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00006_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:13,791 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:13,855 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:15,884 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:16,878 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00007_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:16,886 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:16,950 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:19,119 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:20,116 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00008_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:20,125 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:20,188 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:22,244 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:23,243 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00009_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:23,252 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:23,316 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:25,456 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:26,456 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00010_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:26,464 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:26,528 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:28,574 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:29,570 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00011_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:29,579 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:29,642 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:31,726 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:32,720 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00012_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:32,728 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:32,791 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:34,854 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:35,865 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00013_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:35,873 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:35,936 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:37,971 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:38,962 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00014_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:38,970 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:39,033 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:41,018 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:42,009 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00015_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:42,017 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:42,081 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:44,139 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:45,138 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00016_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:45,146 - stpipe.Image3Pipeline.outlier_detection - INFO - 1 exposures to drizzle together
2024-12-10 20:30:45,209 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:30:47,233 - stpipe.Image3Pipeline.outlier_detection - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:30:48,229 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00017_nis_a3001_outlier_i2d.fits
2024-12-10 20:30:48,235 - stpipe.Image3Pipeline.outlier_detection - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/jwst/associations/association.py:215: UserWarning: Input association file contains path information; note that this can complicate usage and/or sharing of such files.
warnings.warn(err_str, UserWarning)
2024-12-10 20:31:07,749 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00001_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,767 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00002_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,781 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00003_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,795 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00004_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,809 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00005_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,823 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00006_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,838 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00007_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,855 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00008_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,872 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00009_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,889 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00010_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,903 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00011_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,917 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00012_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,931 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00013_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,949 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00014_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,969 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00015_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:07,988 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00016_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:08,005 - stpipe.Image3Pipeline.outlier_detection - INFO - Removing file image3/jw01475006001_02201_00017_nis_a3001_outlier_i2d.fits
2024-12-10 20:31:10,015 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:10,266 - stpipe.Image3Pipeline.outlier_detection - INFO - 4019 pixels marked as outliers
2024-12-10 20:31:12,322 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:12,572 - stpipe.Image3Pipeline.outlier_detection - INFO - 3771 pixels marked as outliers
2024-12-10 20:31:14,650 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:14,897 - stpipe.Image3Pipeline.outlier_detection - INFO - 3756 pixels marked as outliers
2024-12-10 20:31:16,959 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:17,220 - stpipe.Image3Pipeline.outlier_detection - INFO - 4370 pixels marked as outliers
2024-12-10 20:31:19,305 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:19,556 - stpipe.Image3Pipeline.outlier_detection - INFO - 4129 pixels marked as outliers
2024-12-10 20:31:21,623 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:21,874 - stpipe.Image3Pipeline.outlier_detection - INFO - 3899 pixels marked as outliers
2024-12-10 20:31:23,916 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:24,163 - stpipe.Image3Pipeline.outlier_detection - INFO - 3671 pixels marked as outliers
2024-12-10 20:31:26,188 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:26,436 - stpipe.Image3Pipeline.outlier_detection - INFO - 4070 pixels marked as outliers
2024-12-10 20:31:28,458 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:28,704 - stpipe.Image3Pipeline.outlier_detection - INFO - 3982 pixels marked as outliers
2024-12-10 20:31:30,751 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:30,999 - stpipe.Image3Pipeline.outlier_detection - INFO - 4062 pixels marked as outliers
2024-12-10 20:31:33,044 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:33,292 - stpipe.Image3Pipeline.outlier_detection - INFO - 4205 pixels marked as outliers
2024-12-10 20:31:35,330 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:35,578 - stpipe.Image3Pipeline.outlier_detection - INFO - 3822 pixels marked as outliers
2024-12-10 20:31:37,608 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:37,855 - stpipe.Image3Pipeline.outlier_detection - INFO - 3900 pixels marked as outliers
2024-12-10 20:31:39,882 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:40,128 - stpipe.Image3Pipeline.outlier_detection - INFO - 4087 pixels marked as outliers
2024-12-10 20:31:42,166 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:42,413 - stpipe.Image3Pipeline.outlier_detection - INFO - 4027 pixels marked as outliers
2024-12-10 20:31:44,451 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:44,708 - stpipe.Image3Pipeline.outlier_detection - INFO - 3969 pixels marked as outliers
2024-12-10 20:31:46,763 - stpipe.Image3Pipeline.outlier_detection - INFO - Blotting (2048, 2048) <-- (2568, 2545)
2024-12-10 20:31:47,009 - stpipe.Image3Pipeline.outlier_detection - INFO - 3917 pixels marked as outliers
2024-12-10 20:31:47,324 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00001_nis_a3001_crf.fits
2024-12-10 20:31:47,590 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00002_nis_a3001_crf.fits
2024-12-10 20:31:47,855 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00003_nis_a3001_crf.fits
2024-12-10 20:31:48,119 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00004_nis_a3001_crf.fits
2024-12-10 20:31:48,384 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00005_nis_a3001_crf.fits
2024-12-10 20:31:48,654 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00006_nis_a3001_crf.fits
2024-12-10 20:31:48,919 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00007_nis_a3001_crf.fits
2024-12-10 20:31:49,181 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00008_nis_a3001_crf.fits
2024-12-10 20:31:49,446 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00009_nis_a3001_crf.fits
2024-12-10 20:31:49,713 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00010_nis_a3001_crf.fits
2024-12-10 20:31:49,980 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00011_nis_a3001_crf.fits
2024-12-10 20:31:50,244 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00012_nis_a3001_crf.fits
2024-12-10 20:31:50,509 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00013_nis_a3001_crf.fits
2024-12-10 20:31:50,777 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00014_nis_a3001_crf.fits
2024-12-10 20:31:51,299 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00015_nis_a3001_crf.fits
2024-12-10 20:31:51,563 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00016_nis_a3001_crf.fits
2024-12-10 20:31:51,829 - stpipe.Image3Pipeline.outlier_detection - INFO - Saved model in image3/jw01475006001_02201_00017_nis_a3001_crf.fits
2024-12-10 20:31:51,829 - stpipe.Image3Pipeline.outlier_detection - INFO - Step outlier_detection done
2024-12-10 20:31:52,058 - stpipe.Image3Pipeline.resample - INFO - Step resample running with args (<jwst.datamodels.library.ModelLibrary object at 0x7f1ec88e4290>,).
2024-12-10 20:31:52,501 - stpipe.Image3Pipeline.resample - INFO - Driz parameter kernel: square
2024-12-10 20:31:52,501 - stpipe.Image3Pipeline.resample - INFO - Driz parameter pixfrac: 1.0
2024-12-10 20:31:52,502 - stpipe.Image3Pipeline.resample - INFO - Driz parameter fillval: NAN
2024-12-10 20:31:52,502 - stpipe.Image3Pipeline.resample - INFO - Driz parameter weight_type: ivm
2024-12-10 20:31:52,503 - stpipe.Image3Pipeline.resample - INFO - Output pixel scale ratio: 1.0
2024-12-10 20:31:52,646 - stpipe.Image3Pipeline.resample - INFO - Computed output pixel scale: 0.06556276592712629 arcsec.
2024-12-10 20:31:52,716 - stpipe.Image3Pipeline.resample - INFO - Resampling science data
2024-12-10 20:31:52,788 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:31:54,738 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:31:55,622 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:31:57,580 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:31:58,471 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:00,427 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:01,319 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:03,312 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:04,203 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:06,199 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:07,090 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:09,052 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:09,941 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:11,889 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:12,783 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:14,746 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:15,637 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:17,593 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:18,483 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:20,473 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:21,364 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:23,384 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:24,293 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:26,247 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:27,140 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:29,100 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:29,994 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:31,960 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:32,849 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:34,807 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:35,704 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:37,666 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:38,559 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:40,540 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:41,775 - stpipe.Image3Pipeline.resample - INFO - Resampling variance components
2024-12-10 20:32:41,821 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:44,070 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:44,997 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:47,071 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:47,985 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:50,011 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:50,918 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:52,913 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:53,830 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:55,865 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:56,767 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:32:58,820 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:32:59,718 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:01,695 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:02,622 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:04,629 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:05,531 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:07,511 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:08,414 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:10,377 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:11,291 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:13,304 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:14,214 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:16,238 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:17,152 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:19,158 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:20,083 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:22,039 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:22,940 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:24,894 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:25,791 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:27,762 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:28,672 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:30,632 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:31,534 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:33,504 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:34,410 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:36,394 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:37,306 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:39,277 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:40,182 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:42,165 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:43,072 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:45,056 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:45,979 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:47,936 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:48,844 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:50,808 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:51,720 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:53,690 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:54,609 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:56,557 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:33:57,456 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:33:59,394 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:00,295 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:02,270 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:03,178 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:05,155 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:06,054 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:08,016 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:08,920 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:10,880 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:11,792 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:13,761 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:14,663 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:16,659 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:17,559 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:19,557 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:20,480 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:22,447 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:23,348 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:25,323 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:26,221 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:28,180 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:29,088 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:31,092 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:31,990 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:33,943 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:34,850 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:36,829 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:37,753 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:39,739 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:40,640 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:42,633 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:43,532 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:45,513 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:46,424 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:48,391 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:49,296 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:51,313 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:52,224 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:54,209 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:55,123 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:34:57,110 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:34:58,016 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:35:00,024 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:35:00,933 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:35:02,938 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:35:03,861 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:35:05,846 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:35:06,757 - stpipe.Image3Pipeline.resample - WARNING - /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/drizzle/util.py:28: DeprecationWarning: 'is_blank()' has been deprecated since version 2.0.0 and it will be removed in a future release. Please replace calls to 'is_blank()' with alternative implementation.
warnings.warn(
2024-12-10 20:35:08,716 - stpipe.Image3Pipeline.resample - INFO - Drizzling (2048, 2048) --> (2568, 2545)
2024-12-10 20:35:09,663 - stpipe.Image3Pipeline.resample - INFO - Update S_REGION to POLYGON ICRS 303.737924213 -26.798107375 303.788749306 -26.786752702 303.776138546 -26.741791949 303.725332311 -26.753142128
2024-12-10 20:35:10,257 - stpipe.Image3Pipeline.resample - INFO - Saved model in image3/image3_association_i2d.fits
2024-12-10 20:35:10,257 - stpipe.Image3Pipeline.resample - INFO - Step resample done
2024-12-10 20:35:10,498 - stpipe.Image3Pipeline.source_catalog - INFO - Step source_catalog running with args (<ImageModel(2568, 2545) from image3_association_i2d.fits>,).
2024-12-10 20:35:10,514 - stpipe.Image3Pipeline.source_catalog - INFO - Using APCORR reference file: crds_cache/references/jwst/niriss/jwst_niriss_apcorr_0008.fits
2024-12-10 20:35:10,522 - stpipe.Image3Pipeline.source_catalog - INFO - Using ABVEGAOFFSET reference file: crds_cache/references/jwst/niriss/jwst_niriss_abvegaoffset_0003.asdf
2024-12-10 20:35:10,523 - stpipe.Image3Pipeline.source_catalog - INFO - Instrument: NIRISS
2024-12-10 20:35:10,523 - stpipe.Image3Pipeline.source_catalog - INFO - Detector: NIS
2024-12-10 20:35:10,524 - stpipe.Image3Pipeline.source_catalog - INFO - Filter: CLEAR
2024-12-10 20:35:10,524 - stpipe.Image3Pipeline.source_catalog - INFO - Pupil: F150W
2024-12-10 20:35:10,525 - stpipe.Image3Pipeline.source_catalog - INFO - Subarray: FULL
2024-12-10 20:35:10,568 - stpipe.Image3Pipeline.source_catalog - INFO - AB to Vega magnitude offset 1.19753
2024-12-10 20:35:15,004 - stpipe.Image3Pipeline.source_catalog - INFO - Detected 2389 sources
2024-12-10 20:35:16,599 - stpipe.Image3Pipeline.source_catalog - INFO - Wrote source catalog: image3/image3_association_cat.ecsv
2024-12-10 20:35:16,748 - stpipe.Image3Pipeline.source_catalog - INFO - Saved model in image3/image3_association_segm.fits
2024-12-10 20:35:16,749 - stpipe.Image3Pipeline.source_catalog - INFO - Wrote segmentation map: image3_association_segm.fits
2024-12-10 20:35:16,751 - stpipe.Image3Pipeline.source_catalog - INFO - Step source_catalog done
2024-12-10 20:35:16,779 - stpipe.Image3Pipeline - INFO - Step Image3Pipeline done
2024-12-10 20:35:16,779 - stpipe - INFO - Results used jwst version: 1.16.1
Verify which pipeline steps were run#
# Identify *_i2d file and open as datamodel
i2d = glob.glob(os.path.join(image3_dir, "*_i2d.fits"))[0]
i2d_f = datamodels.open(i2d)
i2d_f.meta.cal_step.instance
{'assign_wcs': 'COMPLETE',
'charge_migration': 'COMPLETE',
'clean_flicker_noise': 'SKIPPED',
'dark_sub': 'COMPLETE',
'dq_init': 'COMPLETE',
'flat_field': 'COMPLETE',
'gain_scale': 'SKIPPED',
'group_scale': 'SKIPPED',
'ipc': 'SKIPPED',
'jump': 'COMPLETE',
'linearity': 'COMPLETE',
'outlier_detection': 'COMPLETE',
'persistence': 'SKIPPED',
'photom': 'COMPLETE',
'ramp_fit': 'COMPLETE',
'refpix': 'COMPLETE',
'resample': 'COMPLETE',
'saturation': 'COMPLETE',
'skymatch': 'COMPLETE',
'superbias': 'COMPLETE',
'tweakreg': 'COMPLETE'}
Check which reference files were used to calibrate the dataset:
i2d_f.meta.ref_file.instance
{'area': {'name': 'crds://jwst_niriss_area_0021.fits'},
'camera': {'name': 'N/A'},
'collimator': {'name': 'N/A'},
'crds': {'context_used': 'jwst_1303.pmap', 'sw_version': '12.0.8'},
'dark': {'name': 'crds://jwst_niriss_dark_0169.fits'},
'dflat': {'name': 'N/A'},
'disperser': {'name': 'N/A'},
'distortion': {'name': 'crds://jwst_niriss_distortion_0047.asdf'},
'fflat': {'name': 'N/A'},
'filteroffset': {'name': 'crds://jwst_niriss_filteroffset_0006.asdf'},
'flat': {'name': 'crds://jwst_niriss_flat_0282.fits'},
'fore': {'name': 'N/A'},
'fpa': {'name': 'N/A'},
'gain': {'name': 'crds://jwst_niriss_gain_0006.fits'},
'ifufore': {'name': 'N/A'},
'ifupost': {'name': 'N/A'},
'ifuslicer': {'name': 'N/A'},
'linearity': {'name': 'crds://jwst_niriss_linearity_0017.fits'},
'mask': {'name': 'crds://jwst_niriss_mask_0017.fits'},
'msa': {'name': 'N/A'},
'ote': {'name': 'N/A'},
'photom': {'name': 'crds://jwst_niriss_photom_0043.fits'},
'readnoise': {'name': 'crds://jwst_niriss_readnoise_0005.fits'},
'regions': {'name': 'N/A'},
'saturation': {'name': 'crds://jwst_niriss_saturation_0015.fits'},
'sflat': {'name': 'N/A'},
'specwcs': {'name': 'N/A'},
'superbias': {'name': 'crds://jwst_niriss_superbias_0183.fits'},
'wavelengthrange': {'name': 'N/A'}}
Display combined image#
Visualize the drizzle-combined image.
# Create an Imviz instance and set up default viewer
imviz_i2d = Imviz()
viewer_i2d = imviz_i2d.default_viewer
# Read in the science array for our visualization dataset:
i2d_science = i2d_f.data
# Load the dataset into Imviz
imviz_i2d.load_data(i2d_science)
# Visualize the dataset:
imviz_i2d.show()

plotopt = imviz_i2d.plugins['Plot Options']
plotopt.stretch_function = 'sqrt'
plotopt.image_colormap = 'Viridis'
plotopt.stretch_preset = '95%'
plotopt.zoom_radius = 1024
The viewer looks like this:
viewer_i2d.save('./i2d_science.png')
Image('./i2d_science.png')
Visualize Detected Sources#
Using the source catalog created by the IMAGE3 stage of the pipeline, mark the detected sources, using different markers for point sources and extended sources. The source catalog is saved in image3/image3_association_cat.ecsv file. We will need to read in the i2d file again to make sure the world coordinate system (WCS) info is read in.
Read in catalog file and identify point/extended sources#
catalog_file = glob.glob(os.path.join(image3_dir, "*_cat.ecsv"))[0]
catalog = Table.read(catalog_file)
# To identify point/extended sources, use the 'is_extended' column in the source catalog
pt_src, = np.where(~catalog['is_extended'])
ext_src, = np.where(catalog['is_extended'])
# Define coordinates of point and extended sources
pt_coord = Table({'coord': [SkyCoord(ra=catalog['sky_centroid'][pt_src].ra,
dec=catalog['sky_centroid'][pt_src].dec)]})
ext_coord = Table({'coord': [SkyCoord(ra=catalog['sky_centroid'][ext_src].ra,
dec=catalog['sky_centroid'][ext_src].dec)]})
Mark the extended and point sources on the image#
Display combined image:
# Read in i2d file to Imviz
imviz_cat = Imviz()
viewer_cat = imviz_cat.default_viewer
imviz_cat.load_data(i2d)
imviz_cat.show()

# Adjust settings for viewer
plotopt = imviz_cat.plugins['Plot Options']
plotopt.stretch_function = 'sqrt'
plotopt.image_colormap = 'Viridis'
plotopt.stretch_preset = '95%'
plotopt.zoom_radius = 1024
Point sources will be marked by small pink circles and extended sources will be marked by larger white circles.
# Add marker for point sources:
viewer_cat.marker = {'color': 'pink', 'markersize': 50, 'fill': False}
viewer_cat.add_markers(pt_coord, use_skycoord=True, marker_name='point_sources')
# Add marker for extended sources:
viewer_cat.marker = {'color': 'white', 'markersize': 100, 'fill': False}
viewer_cat.add_markers(ext_coord, use_skycoord=True, marker_name='extended_sources')
Viewer looks like this:
viewer_cat.save('./i2d_markers.png')
Image('./i2d_markers.png')
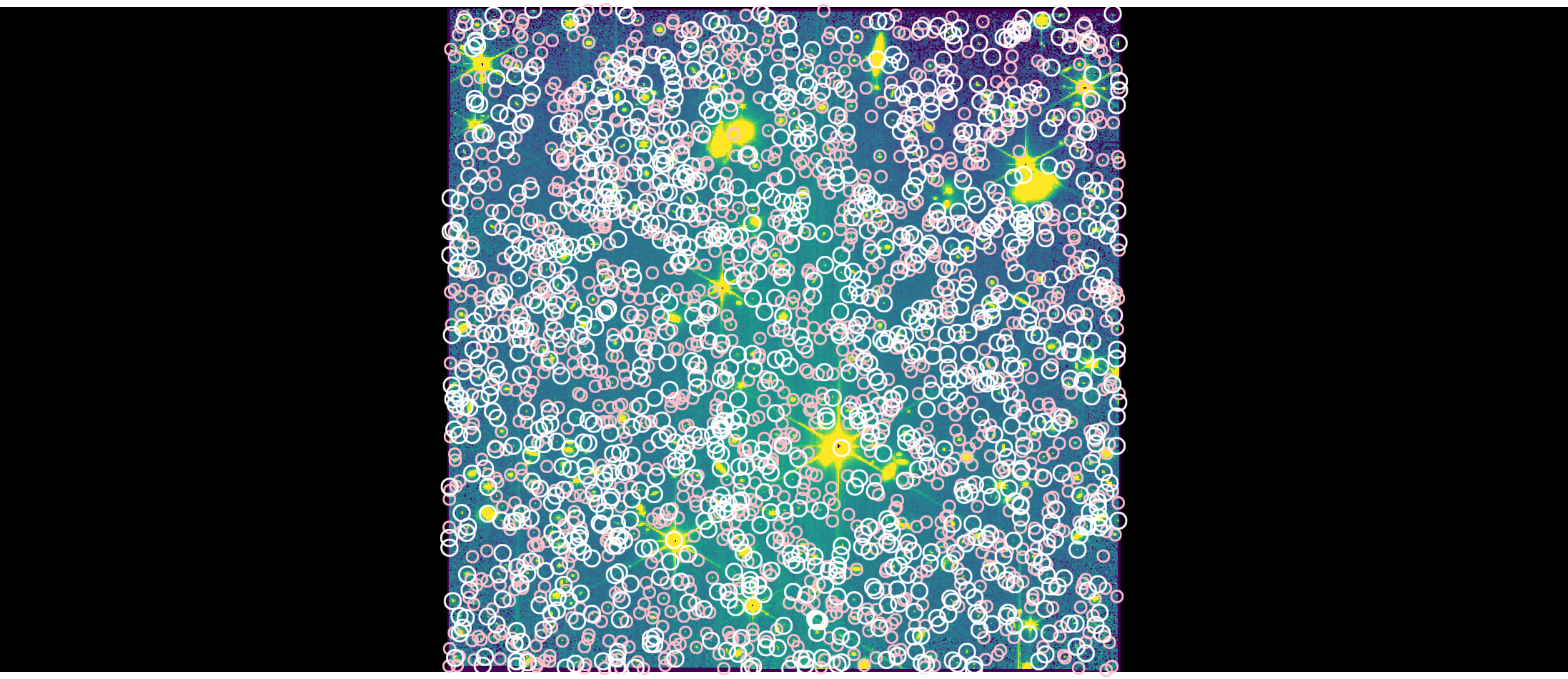
From zoooming in on different portions of the image, we can see that some saturated point sources are erroneously labeled as point sources, some extended and saturated objects are not detected, parts of diffraction spikes are sometimes detected as “extended sources”, and in some cases, the detected source centroid is offset from the center of a source. It is recommended for users to optimize source detection for their science goals using either their own tools or by updating parameters to the source_catalog step of the pipeline.
