
Correcting for Missing Wavecals with Cross-Correlation#
Table of Contents#
1. Introduction#
If the wavelength calibration fails due to, for example, a bad acquisition, the zero point in the spectral direction of the spectrum might be shifted because of the imprecise target positioning. However, if the target was observed multiple times and at least one has the correct zero point, then this shift can be corrected using cross-correlation. In this notebook, we will go through how to fix the shifted spectrum by cross-correlating it with a calibrated one.
1.1 Import Necessary Packages#
astropy.io fitsastropy.table Tablefor accessing FITS filesastroquery.mast Observationsfor finding and downloading data from the MAST archiveastropy.modeling fittingastropy.modeling.models Polynomial1Dfor fitting polynomialsscipy.signal correlatefor performing cross-correlationmatplotlibfor plotting datanumpyfor handling array functionsstistoolsfor quick operations on STIS Dataos,shutil,pathlibfor managing system paths
from astropy.io import fits
from astroquery.mast import Observations
from astropy.modeling import fitting
from astropy.modeling.models import Polynomial1D
from scipy.signal import correlate
from scipy.signal import correlation_lags
import matplotlib.pyplot as plt
import numpy as np
import os
import shutil
from pathlib import Path
import stistools
The following tasks in the stistools package can be run with TEAL:
basic2d calstis ocrreject wavecal x1d x2d
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/nmpfit.py:8: UserWarning: NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.
warnings.warn("NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.")
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/gfit.py:18: UserWarning: GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it.Use astropy.modeling instead.
warnings.warn("GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it."
1.2 Collect Data Set From the MAST Archive Using Astroquery#
In this notebook, we need to download two datasets and explore their correlation.
%%capture --no-stderr output
# remove downlaod directory if it already exists
if os.path.exists("./mastDownload"):
shutil.rmtree("./mastDownload")
# Search target object by obs_id
target_id = "odj101050"
ref_id = "odj101060"
target = Observations.query_criteria(obs_id=[target_id, ref_id])
# get a list of files assiciated with that target
target_list = Observations.get_product_list(target)
# Download only the SCIENCE fits files
Observations.download_products(target_list, productType="SCIENCE")
2. _x1d Spectra of the Observations#
2.1 Creating Shifted Spectrum#
In this notebook, we select two datasets of observations (odj101050, odj101060) with the same target (-PHI-LEO), detector (FUV-MAMA), and grating (G140M). We artificially shift one of the spectra (odj101050) to simulate the wavelength zeropoint shifted spectrum due to target acquisition failures, and use the other spectrum (odj101060) as the reference to conduct cross-correlation. To shift the spectrum, we set the “WAVECORR” calibration switch in the raw fits file to “OMIT” and recalibrate the spectrum using Calstis. “WAVECORR” is the calibration step that determines the shift of the image on the detector along each axis, and therefore by turning off the “WAVECORR” calibration switch, wavecal is not performed and the spectrum is systemetically shifted. We will then use the shifted spectrum and the reference spectrum to determine the wavelength zero point shift, recalibrate this shifted spectrum, and compare it with the original pipeline spectrum.
Next, use the Calibration Reference Data System (CRDS) command line tools to update and download the reference files.
crds_path = os.path.expanduser("~") + "/crds_cache"
os.environ["CRDS_PATH"] = crds_path
os.environ["CRDS_SERVER_URL"] = "https://hst-crds.stsci.edu"
os.environ["oref"] = os.path.join(crds_path, "references/hst/oref/")
%%capture --no-stderr output
!crds bestrefs --update-bestrefs --sync-references=1 --files ./mastDownload/HST/odj101050/odj101050_raw.fits
pip_raw = os.path.join("./mastDownload/HST", "{}".format(target_id), "{}_raw.fits".format(target_id))
# Set the "WAVECORR" switch in the raw fits file header to "OMIT"
fits.setval(pip_raw, "WAVECORR", value="OMIT")
# Create and clean "./Shifted" directory for saving new files
shifted_dir = Path("./Shifted")
if os.path.exists(shifted_dir):
shutil.rmtree(shifted_dir)
Path(shifted_dir).mkdir(exist_ok=True)
%%capture --no-stderr output
# Recalibration
res = stistools.calstis.calstis(pip_raw, verbose=False, outroot="./Shifted/")
# calstis returns 0 if calibration completes; if not, raise assertion error
assert res == 0, f"CalSTIS exited with an error: {res}"
*** CALSTIS-0 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:04 UTC
Input ./mastDownload/HST/odj101050/odj101050_raw.fits
Outroot ./Shifted/odj101050_raw.fits
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:04 UTC
Input ./mastDownload/HST/odj101050/odj101050_raw.fits
Output ./Shifted/odj101050_flt.fits
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
Imset 1 Begin 20:16:04 UTC
DQICORR PERFORM
DQITAB oref$uce15153o_bpx.fits
DQITAB PEDIGREE=GROUND
DQITAB DESCRIP =New BPIXTAB with opt_elem column and correct repeller wire flag----
DQICORR COMPLETE
Uncertainty array initialized.
LORSCORR PERFORM
LORSCORR COMPLETE
GLINCORR PERFORM
LFLGCORR PERFORM
MLINTAB oref$j9r16559o_lin.fits
MLINTAB PEDIGREE=GROUND
MLINTAB DESCRIP =T. Danks gathered Info
MLINTAB DESCRIP =T. Danks Gathered Info
GLINCORR COMPLETE
LFLGCORR COMPLETE
DARKCORR PERFORM
DARKFILE oref$q591955ro_drk.fits
DARKFILE PEDIGREE=INFLIGHT 29/01/2003 03/08/2004
DARKFILE DESCRIP =Avg of 152 1380s darks from 9615 10034
DARKCORR COMPLETE
FLATCORR PERFORM
PFLTFILE oref$mbj1658bo_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 17/03/1998 17/05/2002
PFLTFILE DESCRIP =On-orbit FUV P-flat created from proposals 7645,8428,8862,8922
LFLTFILE oref$m1b2139no_lfl.fits
LFLTFILE PEDIGREE=DUMMY
LFLTFILE DESCRIP =Dummy file created by M. Mcgrath. Modified by C Proffitt
FLATCORR COMPLETE
PHOTCORR OMIT
DOPPCORR applied to DQICORR, DARKCORR, FLATCORR
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 1 End 20:16:05 UTC
End 02-Dec-2025 20:16:05 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-7 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:05 UTC
Input ./Shifted/odj101050_flt.fits
Output ./Shifted/odj101050_x2d.fits
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
Imset 1 Begin 20:16:05 UTC
Warning Wavecal processing has not been performed.
HELCORR PERFORM
HELCORR COMPLETE
Order 1 Begin 20:16:05 UTC
X2DCORR PERFORM
DISPCORR PERFORM
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
SDCTAB oref$16j1600ao_sdc.fits
SDCTAB PEDIGREE=INFLIGHT 13/07/1997 13/06/2017
SDCTAB DESCRIP =Co-aligned fiducial bars via an update to the CDELT2 plate scales.-
SDCTAB DESCRIP =updated CDELT2 with inflight data, others Lindler-prelaunch
DISPTAB oref$m7p16110o_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 04/03/1999 04/03/1999
DISPTAB DESCRIP =Dispersion coefficients reference table
DISPTAB DESCRIP =INFLIGHT Cal. Disp. Coeffs
INANGTAB oref$h1v1541eo_iac.fits
INANGTAB PEDIGREE=GROUND
INANGTAB DESCRIP =Prelaunch Calibration/Lindler and Models
INANGTAB DESCRIP =Model
SPTRCTAB oref$77o1827do_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 27/02/1997 30/06/1998
SPTRCTAB DESCRIP =New traces for select echelle secondary modes
SPTRCTAB DESCRIP =Sandoval, Initial Postlaunch Calibration
FLUXCORR PERFORM
PHOTTAB oref$9511857bo_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 28/02/2000 21/11/2001
PHOTTAB DESCRIP =Fullerton 2025 calibration
PHOTTAB DESCRIP =Updated Sensitivities 2025 (CALSPEC v11 models)
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$y2r16005o_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/5/1997 19/12/1998
PCTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
PCTAB DESCRIP = 2 STIS observations averaged
TDSTAB oref$93q1947bo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/02/2025
TDSTAB DESCRIP =Updated time and slope arrays for all optical elements.
FLUXCORR COMPLETE
X2DCORR COMPLETE
DISPCORR COMPLETE
Order 1 End 20:16:05 UTC
Imset 1 End 20:16:05 UTC
End 02-Dec-2025 20:16:05 UTC
*** CALSTIS-7 complete ***
*** CALSTIS-6 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:05 UTC
Warning Grating-aperture throughput correction table (GACTAB) was not found,
and no gac corrections will be applied
Input ./Shifted/odj101050_flt.fits
Output ./Shifted/odj101050_x1d.fits
Rootname odj101050
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
XTRACTAB oref$n7p10323o_1dx.fits
XTRACTAB PEDIGREE=INFLIGHT 29/05/97
XTRACTAB DESCRIP =Analysis from prop. 7064 and ground data
XTRACTAB DESCRIP =Analysis from prop. 7064
SPTRCTAB oref$77o1827do_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 27/02/1997 01/12/2009
SPTRCTAB DESCRIP =New traces for select echelle secondary modes
Imset 1 Begin 20:16:05 UTC
Input read into memory.
Order 1 Begin 20:16:05 UTC
X1DCORR PERFORM
BACKCORR PERFORM
******** Calling Slfit ***********BACKCORR COMPLETE
X1DCORR COMPLETE
Spectrum extracted at y position = 390.008
DISPCORR PERFORM
DISPTAB oref$m7p16110o_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 04/03/1999 04/03/1999
DISPTAB DESCRIP =Dispersion coefficients reference table
DISPTAB DESCRIP =INFLIGHT Cal. Disp. Coeffs
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
INANGTAB oref$h1v1541eo_iac.fits
INANGTAB DESCRIP =Prelaunch Calibration/Lindler and Models
DISPCORR COMPLETE
HELCORR PERFORM
HELCORR COMPLETE
PHOTTAB oref$9511857bo_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 27/02/1997 24/09/2019
PHOTTAB DESCRIP =Fullerton 2025 calibration
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$y2r16005o_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/05/1997 19/12/1998
PCTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
TDSTAB oref$93q1947bo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/02/2025
TDSTAB DESCRIP =Updated time and slope arrays for all optical elements.
FLUXCORR PERFORM
FLUXCORR COMPLETE
SGEOCORR OMIT
Row 1 written to disk.
Order 1 End 20:16:05 UTC
Imset 1 End 20:16:05 UTC
Warning Keyword `XTRACALG' is being added to header.
End 02-Dec-2025 20:16:05 UTC
*** CALSTIS-6 complete ***
End 02-Dec-2025 20:16:05 UTC
*** CALSTIS-0 complete ***
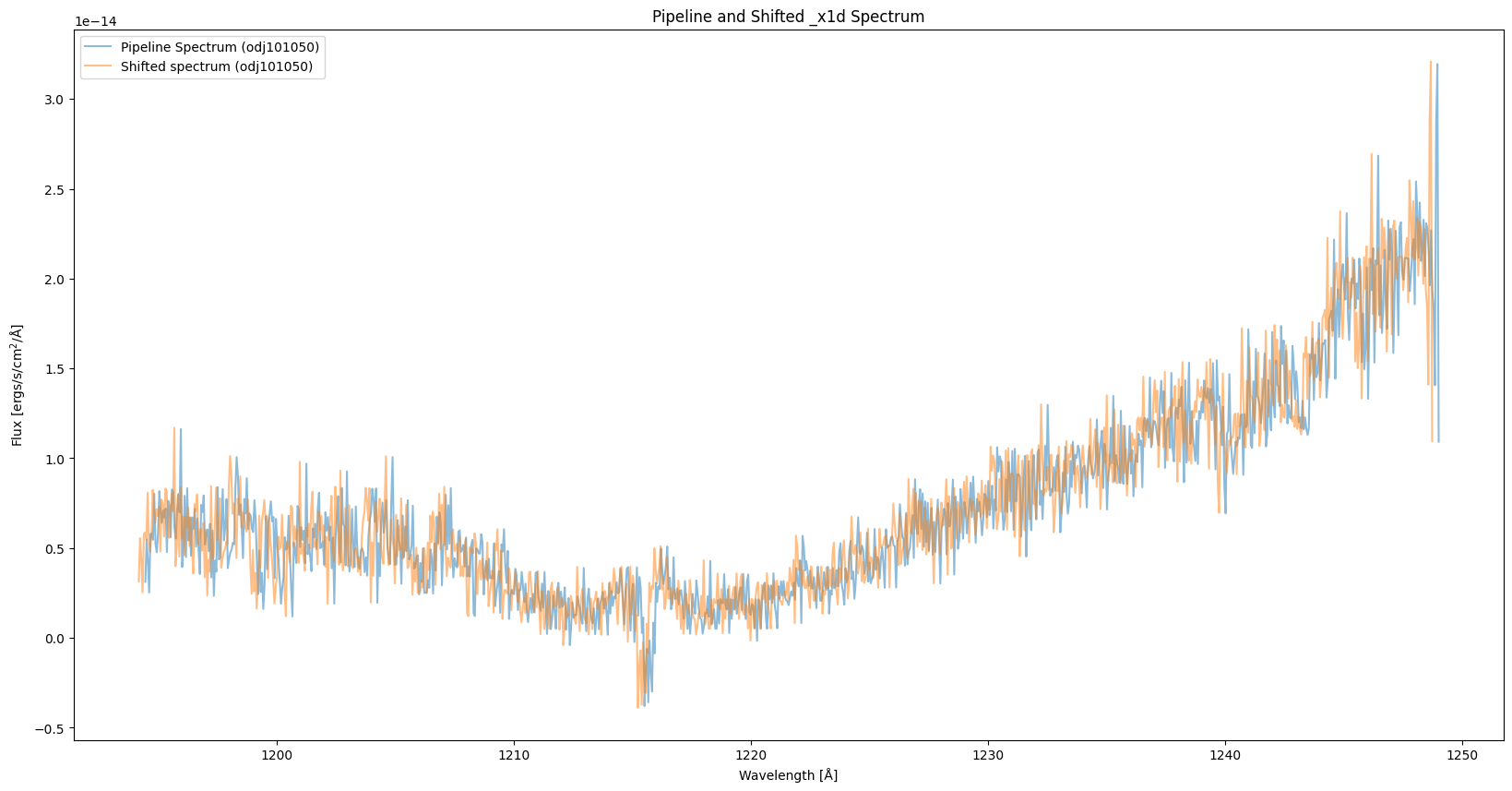
As seen in the plot, the spectrum is now shifted compared to the pipeline spectrum:
pip_x1d = os.path.join("./mastDownload/HST", "{}".format(target_id), "{}_x1d.fits".format(target_id))
shifted_x1d = Path("./Shifted/{}_x1d.fits".format(target_id))
with fits.open(pip_x1d) as hdu1, fits.open(shifted_x1d) as hdu2:
pip_wl = hdu1[1].data["WAVELENGTH"][0]
pip_flux = hdu1[1].data["FLUX"][0]
shifted_wl = hdu2[1].data["WAVELENGTH"][0]
shifted_flux = hdu2[1].data["FLUX"][0]
fig = plt.figure(figsize=(20, 10))
plt.plot(pip_wl, pip_flux, label="Pipeline Spectrum ({})".format(target_id), alpha=0.5)
plt.plot(shifted_wl, shifted_flux, label="Shifted spectrum ({})".format(target_id), alpha=0.5)
plt.legend(loc="best")
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Pipeline and Shifted _x1d Spectrum")
Text(0.5, 1.0, 'Pipeline and Shifted _x1d Spectrum')
2.2 Spectrum Interpolation#
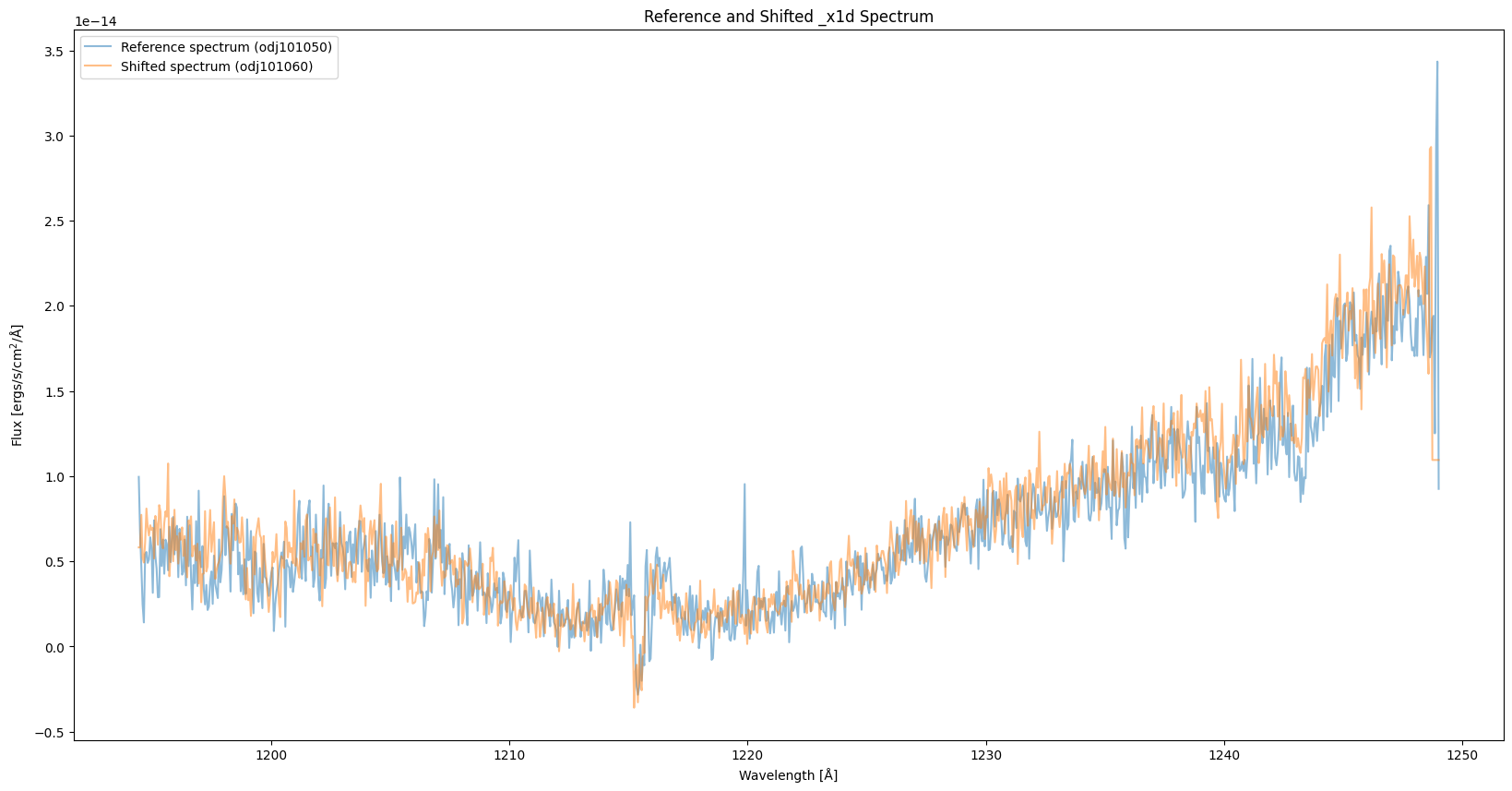
Since the wavelength range of the pipeline and shifted spectra are different, we interpolate one of the spectra based on the wavelength of the other one so that the two spectra share the same wavelength array. As shown in the plot, the spectrum with “WAVECORR” turned off is systemetically shifted to the left.
ref_x1d = os.path.join("./mastDownload/HST", "{}".format(ref_id), "{}_x1d.fits".format(ref_id))
with fits.open(ref_x1d) as hdu1, fits.open(shifted_x1d) as hdu2:
wl = hdu1[1].data["WAVELENGTH"][0]
ref_flux = hdu1[1].data["FLUX"][0]
shifted_wl = hdu2[1].data["WAVELENGTH"][0]
shifted_flux = hdu2[1].data["FLUX"][0]
shifted_flux = np.interp(wl, shifted_wl, shifted_flux)
fig = plt.figure(figsize=(20, 10))
plt.plot(wl, ref_flux, alpha=0.5, label="Reference spectrum ({})".format(target_id))
plt.plot(wl, shifted_flux, alpha=0.5, label="Shifted spectrum ({})".format(ref_id))
plt.legend(loc="best")
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Reference and Shifted _x1d Spectrum")
Text(0.5, 1.0, 'Reference and Shifted _x1d Spectrum')
3. Cross-Correlation#
3.1 Dispersion per Pixel#
To perform cross-correlation, detemine the shift amount in pixels, and then convert it into wavelength, we first need to determine the dispersion per pixel, i.e., the mean differences of adjacent data points in the wavelength grid.
mean_plate_scale = np.mean(wl[1:]-wl[:-1])
mean_plate_scale
print("The dispersion per pixel is {:.3f}".format(mean_plate_scale) + " Å/pixel")
The dispersion per pixel is 0.053 Å/pixel
3.2 Masking out the Lyman-alpha line#
The absorption line at around 1215 Å is from Hydrogen Lyman-alpha, which mostly comes from the atmosphere and so should not shift like the science spectrum. Therefore, we need to mask out this region by separating the spectrum into two parts and perform two cross-correlations. There are other airglows lines in the ultraviolet that also does not shift with the science spectrum, including OI line at 1302 Å, OI line at 1305 Å, OI line at 1306 Å. For more information on the Airglow, see: AIRGLOW.
# the spectrum on the right of Lyman-alpha
ref_flux1 = ref_flux[wl > 1220]
shifted_flux1 = shifted_flux[wl > 1220]
# the spectrum on the left of Lyman-alpha
ref_flux2 = ref_flux[wl < 1213]
shifted_flux2 = shifted_flux[wl < 1213]
3.3 Lag and Cross-Correlation Coefficient#
The lag is the displacement (in pixels) in the lagged spectrum. If the lag is 0, the spectra are aligned and not shifted.
The cross-correlation coefficient decodes how similar two spectra are. The cross-correlation coefficient takes values from -1 to 1: if it’s positive, the 2 spectra are positively correlated, if it’s negative, the 2 spectra are negatively correlated.
The cross-correlation algorithm shifts one of the input spectra according the the lags, and computes the cross-correlation coefficient for each lag. Then we take the lag with the maximum cross-coefficient and compute the corresponding displacement in wavelength space.
In general, the cross-correlation can be written as:
in which k is the lag, C is the cross-correlation coefficient, and x and y are the input spectra.
Normalization of the input spectra is required to ensure the cross-correlation coefficient is in the [-1,1] range.
def cross_correlate(shifted_flux, ref_flux):
assert len(shifted_flux) == len(ref_flux), "Arrays must be same size"
# Normalize inputs:
shifted_flux = shifted_flux - shifted_flux.mean()
shifted_flux /= shifted_flux.std()
ref_flux = ref_flux - ref_flux.mean()
ref_flux /= ref_flux.std()
# centered at the median of len(a)
lag = correlation_lags(len(shifted_flux), len(ref_flux), mode="same")
# find the cross-correlation coefficient
cc = correlate(shifted_flux, ref_flux, mode="same") / float(len(ref_flux))
return lag, cc
3.4 Polynimial Fitting and Zero Point Shift#
After we get the lag and cross-correlation coefficient, we can determine the zero point shift by finding the lag with the maximum cross-correlation coefficient. However, since we only have discrete pixels shifts, we will fit a quadratic curve near the peak, get a fractional pixel shift, and find the maxima of the quadratic curve as the zero point shift. The zero point shift is shown as the green vertical line in the plot.
In this specific case, we choose the lag from -3 to 3, and fitted a 2 degree polynomial curve around the maximum cross-correlation coefficient to determine the shift in pixel space.The lag range and polynomial fitting is not the single solution that can be applied to all cases of wavelength zero point shifts. Users should experiment with the lag range and number of points to fit the polynomial curve based on the use case.
We first find the lag and cross-correlation coefficient of the right part of the spectrum:
fig = plt.figure(figsize=(10, 6))
lag, cc = cross_correlate(shifted_flux1, ref_flux1)
plt.plot(lag, cc, ".-", label="cross-correlation coefficient")
# fit quadratic near the peak to find the pixel shift
fitter = fitting.LinearLSQFitter()
# get the 5 points near the peak
width = 5
low, hi = np.argmax(cc) - width//2, np.argmax(cc) + width//2 + 1
fit = fitter(Polynomial1D(degree=2), x=lag[low:hi], y=cc[low:hi])
x_c = np.arange(-10, 0, 0.01)
plt.plot(x_c, fit(x_c), alpha=0.5, label="fitted quadratic curve")
# finding the maxima
shift1 = -fit.parameters[1] / (2. * fit.parameters[2])
plt.plot([shift1, shift1], [0, 1], alpha=0.5, label="quadratic curve maxima")
plt.xlim(-20, 20)
plt.ylim(0, 1)
plt.xlabel("Lag [pix]")
plt.ylabel("Cross-correlation coeff")
plt.title("15168-01, G140M/C1222 Observations")
plt.legend(loc="best")
<matplotlib.legend.Legend at 0x7f6696910750>
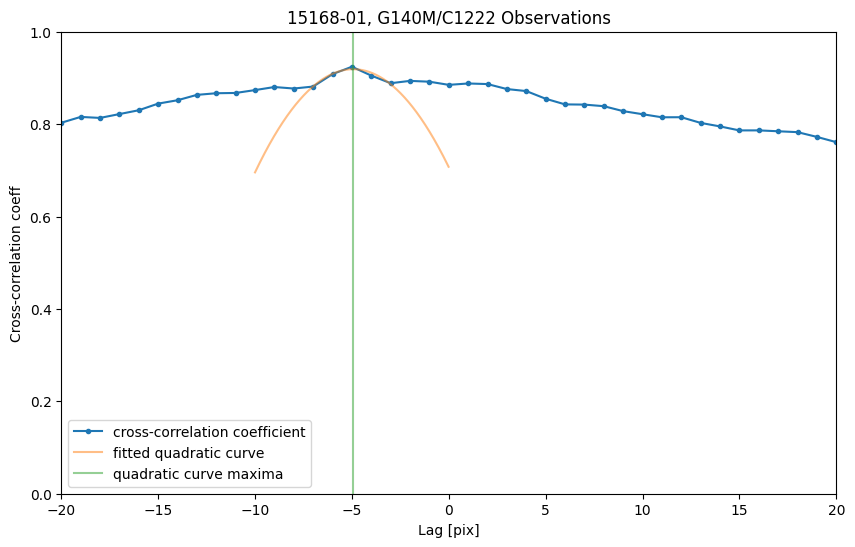
Convert the lag back into zero point shift in wavelength space:
print("Shift between the G140M/c1222 observations is {:.3f} pix = {:.3f}".format(shift1, shift1 * mean_plate_scale) + "Å")
Shift between the G140M/c1222 observations is -4.935 pix = -0.263Å
Apply the same procedure to the left part of the spectrum:
fig = plt.figure(figsize=(10, 6))
lag, cc = cross_correlate(shifted_flux2, ref_flux2)
plt.plot(lag, cc, ".-", label="cross-correlation coefficient")
# fit quadratic near the peak to find the pixel shift
fitter = fitting.LinearLSQFitter()
# get the 5 points near the peak
width = 5
low, hi = np.argmax(cc) - width//2, np.argmax(cc) + width//2 + 1
fit = fitter(Polynomial1D(degree=2), x=lag[low:hi], y=cc[low:hi])
x_c = np.arange(-10, 0, 0.01)
plt.plot(x_c, fit(x_c), alpha=0.5, label="fitted quadratic curve")
# finding the maxima
shift2 = -fit.parameters[1] / (2. * fit.parameters[2])
plt.plot([shift2, shift2], [0, 1], alpha=0.5, label="quadratic curve maxima")
plt.xlim(-20, 20)
plt.ylim(0, 1)
plt.xlabel("Lag [pix]")
plt.ylabel("Cross-correlation coeff")
plt.title("15168-01, G140M/C1222 Observations")
plt.legend(loc="best")
print("shift2 between the G140M/c1222 observations is {:.3f} pix = {:.3f}".format(shift2, shift2 * mean_plate_scale) + "Å")
shift2 between the G140M/c1222 observations is -3.956 pix = -0.211Å
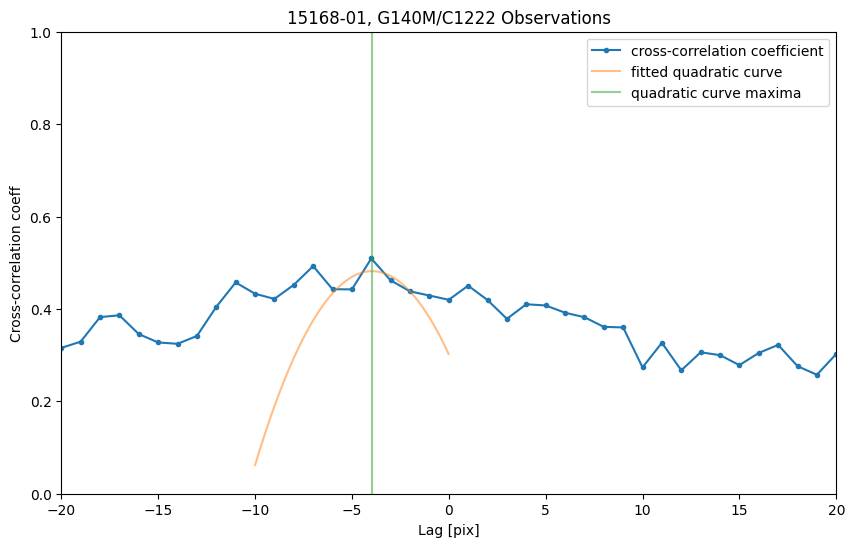
However, as shown in the plot, the maximum cross-correlation coefficient (~0.5) is relatively small, which suggests that the spectra are less similar on the left side. With such a small cross-correlation coefficient, we cannot determine a reasonable shift in the pixel space. Therefore, we only take the shift determined by the right part of the spectrum as the shift of the spectrum:
shift = shift1
print("shift between the G140M/c1222 observations is {:.3f} pix = {:.3f}".format(shift, shift * mean_plate_scale) + "Å")
shift between the G140M/c1222 observations is -4.935 pix = -0.263Å
4. Recalibrate Spectrum#
After we determine the wavelength zero point shift, we can use the value to recalibrate the spectrum.
In the Calstis pipeline, “WAVECORR” calibration step determines the spectral shift values, and writes the keyword values SHIFTA1, SHIFTA2 for the spectral and spatial shifts, respectively, to the science header. To apply the spectral shift from the cross-correlation, we get the SHIFTA1, SHIFTA2 keywords from the _flt fits file, add the shift (in pixel space) to SHIFTA1, and write the updated keywords to the _raw fits file.
# get SHIFTA1, SHIFTA1 keywords from the first science extension
shifted_flt = Path("./Shifted/{}_flt.fits".format(target_id))
# since we have turned off WAVECOR at the beginning, SHIFTA1 should be 0
SHIFTA1 = fits.getval(shifted_flt, "SHIFTA1", 1)
SHIFTA2 = fits.getval(shifted_flt, "SHIFTA2", 1)
assert (SHIFTA1 == 0 and SHIFTA2 == 0)
# update SHIFTA1 (only in the spectral direction)
SHIFTA1 += shift
# update the the SHIFTA1, SHIFTA1 keywords in the _raw fits file first science extension
fits.setval(pip_raw, "SHIFTA1", value=SHIFTA1, ext=1)
fits.setval(pip_raw, "SHIFTA2", value=SHIFTA2, ext=1)
Then we need to turn the “WAVECORR” calibration switch to “omit” so that the Calstis calibration pipeline takes our new computed value instead of the wavecal returned value as the wavelength zero point shift.
fits.setval(pip_raw, "WAVECORR", value="OMIT", ext=0)
assert fits.getval(pip_raw, keyword="WAVECORR", ext=0) == "OMIT"
Recalibrate the spectrum using the Calstis pipeline. We create a new folder called “Recalibration” to store the recalibrated files. For more information on the Calstis pipeline, see: STIS Calibration, calstis.
%%capture --no-stderr output
# Create and clean "./Recalibration" directory for saving new files
if os.path.exists("./Recalibration"):
shutil.rmtree("./Recalibration")
Path("./Recalibration").mkdir(exist_ok=True)
# Recalibration
res = stistools.calstis.calstis(pip_raw, verbose=False, outroot="./Recalibration/")
# calstis returns 0 if calibration completes; if not, raise assertion error
assert res == 0, f"CalSTIS exited with an error: {res}"
recal_x1d = Path("./Recalibration/{}_x1d.fits".format(target_id))
*** CALSTIS-0 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:06 UTC
Input ./mastDownload/HST/odj101050/odj101050_raw.fits
Outroot ./Recalibration/odj101050_raw.fits
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:06 UTC
Input ./mastDownload/HST/odj101050/odj101050_raw.fits
Output ./Recalibration/odj101050_flt.fits
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
Imset 1 Begin 20:16:06 UTC
DQICORR PERFORM
DQITAB oref$uce15153o_bpx.fits
DQITAB PEDIGREE=GROUND
DQITAB DESCRIP =New BPIXTAB with opt_elem column and correct repeller wire flag----
DQICORR COMPLETE
Uncertainty array initialized.
LORSCORR PERFORM
LORSCORR COMPLETE
GLINCORR PERFORM
LFLGCORR PERFORM
MLINTAB oref$j9r16559o_lin.fits
MLINTAB PEDIGREE=GROUND
MLINTAB DESCRIP =T. Danks gathered Info
MLINTAB DESCRIP =T. Danks Gathered Info
GLINCORR COMPLETE
LFLGCORR COMPLETE
DARKCORR PERFORM
DARKFILE oref$q591955ro_drk.fits
DARKFILE PEDIGREE=INFLIGHT 29/01/2003 03/08/2004
DARKFILE DESCRIP =Avg of 152 1380s darks from 9615 10034
DARKCORR COMPLETE
FLATCORR PERFORM
PFLTFILE oref$mbj1658bo_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 17/03/1998 17/05/2002
PFLTFILE DESCRIP =On-orbit FUV P-flat created from proposals 7645,8428,8862,8922
LFLTFILE oref$m1b2139no_lfl.fits
LFLTFILE PEDIGREE=DUMMY
LFLTFILE DESCRIP =Dummy file created by M. Mcgrath. Modified by C Proffitt
FLATCORR COMPLETE
PHOTCORR OMIT
DOPPCORR applied to DQICORR, DARKCORR, FLATCORR
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 1 End 20:16:06 UTC
End 02-Dec-2025 20:16:06 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-7 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:06 UTC
Input ./Recalibration/odj101050_flt.fits
Output ./Recalibration/odj101050_x2d.fits
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
Imset 1 Begin 20:16:06 UTC
Warning Wavecal processing has not been performed.
HELCORR PERFORM
HELCORR COMPLETE
Order 1 Begin 20:16:06 UTC
X2DCORR PERFORM
DISPCORR PERFORM
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
SDCTAB oref$16j1600ao_sdc.fits
SDCTAB PEDIGREE=INFLIGHT 13/07/1997 13/06/2017
SDCTAB DESCRIP =Co-aligned fiducial bars via an update to the CDELT2 plate scales.-
SDCTAB DESCRIP =updated CDELT2 with inflight data, others Lindler-prelaunch
DISPTAB oref$m7p16110o_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 04/03/1999 04/03/1999
DISPTAB DESCRIP =Dispersion coefficients reference table
DISPTAB DESCRIP =INFLIGHT Cal. Disp. Coeffs
INANGTAB oref$h1v1541eo_iac.fits
INANGTAB PEDIGREE=GROUND
INANGTAB DESCRIP =Prelaunch Calibration/Lindler and Models
INANGTAB DESCRIP =Model
SPTRCTAB oref$77o1827do_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 27/02/1997 30/06/1998
SPTRCTAB DESCRIP =New traces for select echelle secondary modes
SPTRCTAB DESCRIP =Sandoval, Initial Postlaunch Calibration
FLUXCORR PERFORM
PHOTTAB oref$9511857bo_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 28/02/2000 21/11/2001
PHOTTAB DESCRIP =Fullerton 2025 calibration
PHOTTAB DESCRIP =Updated Sensitivities 2025 (CALSPEC v11 models)
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$y2r16005o_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/5/1997 19/12/1998
PCTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
PCTAB DESCRIP = 2 STIS observations averaged
TDSTAB oref$93q1947bo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/02/2025
TDSTAB DESCRIP =Updated time and slope arrays for all optical elements.
FLUXCORR COMPLETE
X2DCORR COMPLETE
DISPCORR COMPLETE
Order 1 End 20:16:06 UTC
Imset 1 End 20:16:06 UTC
End 02-Dec-2025 20:16:06 UTC
*** CALSTIS-7 complete ***
*** CALSTIS-6 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:16:06 UTC
Warning Grating-aperture throughput correction table (GACTAB) was not found,
and no gac corrections will be applied
Input ./Recalibration/odj101050_flt.fits
Output ./Recalibration/odj101050_x1d.fits
Rootname odj101050
OBSMODE ACCUM
APERTURE 52X0.2
OPT_ELEM G140M
DETECTOR FUV-MAMA
XTRACTAB oref$n7p10323o_1dx.fits
XTRACTAB PEDIGREE=INFLIGHT 29/05/97
XTRACTAB DESCRIP =Analysis from prop. 7064 and ground data
XTRACTAB DESCRIP =Analysis from prop. 7064
SPTRCTAB oref$77o1827do_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 27/02/1997 01/12/2009
SPTRCTAB DESCRIP =New traces for select echelle secondary modes
Imset 1 Begin 20:16:06 UTC
Input read into memory.
Order 1 Begin 20:16:06 UTC
X1DCORR PERFORM
BACKCORR PERFORM
******** Calling Slfit ***********BACKCORR COMPLETE
X1DCORR COMPLETE
Spectrum extracted at y position = 390.008
DISPCORR PERFORM
DISPTAB oref$m7p16110o_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 04/03/1999 04/03/1999
DISPTAB DESCRIP =Dispersion coefficients reference table
DISPTAB DESCRIP =INFLIGHT Cal. Disp. Coeffs
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
INANGTAB oref$h1v1541eo_iac.fits
INANGTAB DESCRIP =Prelaunch Calibration/Lindler and Models
DISPCORR COMPLETE
HELCORR PERFORM
HELCORR COMPLETE
PHOTTAB oref$9511857bo_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 27/02/1997 24/09/2019
PHOTTAB DESCRIP =Fullerton 2025 calibration
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$y2r16005o_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/05/1997 19/12/1998
PCTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
TDSTAB oref$93q1947bo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/02/2025
TDSTAB DESCRIP =Updated time and slope arrays for all optical elements.
FLUXCORR PERFORM
FLUXCORR COMPLETE
SGEOCORR OMIT
Row 1 written to disk.
Order 1 End 20:16:06 UTC
Imset 1 End 20:16:06 UTC
Warning Keyword `XTRACALG' is being added to header.
End 02-Dec-2025 20:16:06 UTC
*** CALSTIS-6 complete ***
End 02-Dec-2025 20:16:06 UTC
*** CALSTIS-0 complete ***
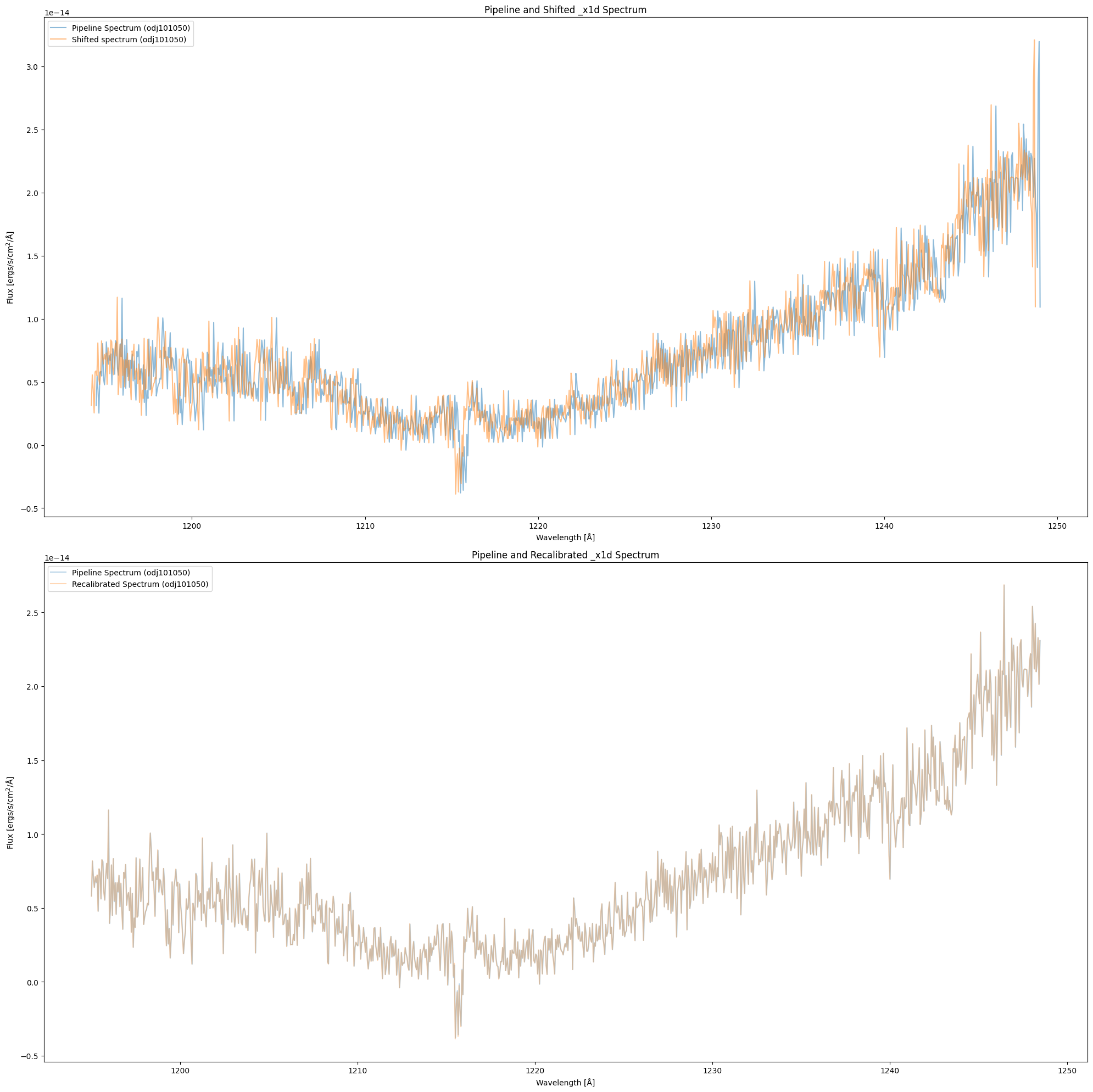
We compare the recalibrated spectrum with the pipeline spectrum. The top panel is the shifted spectrum (orange) and the pipeline spectrum (blue) of observation odj101050, which is the same as the plot in 2.1 Creating Shifted Spectrum. The bottom panel is the recalibrated spectrum (orange) and the pipeline spectrum (blue). The spectra almost overlap in the bottom panel, which suggests that the wavelength shift issue is solved in the recalibrated spectrum.
fig = plt.figure(figsize=(20, 20))
plt.subplot(2, 1, 1)
with fits.open(pip_x1d) as hdu1, fits.open(shifted_x1d) as hdu2:
pip_wl = hdu1[1].data["WAVELENGTH"][0]
pip_flux = hdu1[1].data["FLUX"][0]
shifted_wl = hdu2[1].data["WAVELENGTH"][0]
shifted_flux = hdu2[1].data["FLUX"][0]
plt.plot(pip_wl, pip_flux, label="Pipeline Spectrum ({})".format(target_id), alpha=0.5)
plt.plot(shifted_wl, shifted_flux, label="Shifted spectrum ({})".format(target_id), alpha=0.5)
plt.legend(loc="best")
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Pipeline and Shifted _x1d Spectrum")
plt.subplot(2, 1, 2)
with fits.open(pip_x1d) as hdu1, fits.open(recal_x1d) as hdu2:
wl1 = hdu1[1].data["WAVELENGTH"][0][10:-10]
wl2 = hdu2[1].data["WAVELENGTH"][0][10:-10]
flux1 = hdu1[1].data["FLUX"][0][10:-10]
flux2 = hdu2[1].data["FLUX"][0][10:-10]
plt.plot(wl1, flux1, label="Pipeline Spectrum ({})".format(target_id), alpha=0.3)
plt.plot(wl2, flux2, label="Recalibrated Spectrum ({})".format(target_id), alpha=0.3)
plt.legend(loc="best")
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Pipeline and Recalibrated _x1d Spectrum")
plt.tight_layout()
About this Notebook#
Author: Keyi Ding
Updated On: 2025-11-04
This tutorial was generated to be in compliance with the STScI style guides and would like to cite the Jupyter guide in particular.
Citations#
If you use astropy, matplotlib, astroquery, or numpy for published research, please cite the
authors. Follow these links for more information about citations:


