Aligning HST images to an Absolute Reference Catalog#
Table of Contents#
1. Download the Observations with astroquery
1.1 Check image header data
1.2 Inspect the alignment
2. Query the Reference Catalogs
2.1 Identify Coordinates
2.2 SDSS Query
2.3 Gaia Query
3. Align images with TweakReg
3.1 SDSS Alignment
3.2 Inspect the shift file and fit residuals
3.3 Gaia Alignment
3.4 Inspect the shift file and fit residuals
3.5 Overplot Matched Sources on the image
3.6 Update header with optimal parameters
4. Combine the images using AstroDrizzle
Introduction#
The alignment of HST exposures is a critical step in image stacking or combination performed with software such as AstroDrizzle. Generally, a relative alignment is performed to align one or multiple images to another image that is designated as the reference image. The reference image is generally the deepest exposure and/or that covering the largest area of all the exposures. This process aligns the images to each other, but the pointing error of the observatory can still cause the images to have incorrect absolute astrometry. When absolute astrometry is desired, the images can be aligned to an external catalog with an absolute world coordinate system (WCS). In this example, we will provide a workflow to query catalogs such as SDSS and Gaia using the astroquery package, and then align the images to that catalog using TweakReg.
The workflow in this notebook for aligning images to Gaia is based on WFC3 ISR 2017-19: Aligning HST Images to Gaia: a Faster Mosaicking Workflow and contains a subset of the information and code found in this repository. For more information, see the notebook in that repository titled Gaia_alignment.ipynb.
For more information about TweakReg, see the other notebooks in this repository or the TweakReg Documentation.
For more information on Astroquery, see the other notebooks in this repository or the Astroquery Documentation.
Import Packages#
The following Python packages are required to run the Jupyter Notebook:
os - change and make directories
glob - gather lists of filenames
shutil - remove directories and files
numpy - math and array functions
matplotlib - make figures and graphics
astropy - file handling, tables, units, WCS, statistics
astroquery - download data and query databases
drizzlepac - align and combine HST images
import os
import glob
import shutil
import numpy as np
import matplotlib.pyplot as plt
from collections import defaultdict
from IPython.display import Image, clear_output
import astropy.units as u
from astropy.io import ascii, fits
from astropy.table import Table
from astropy.units import Quantity
from astropy.coordinates import SkyCoord
from astropy.visualization import ZScaleInterval
from astroquery.mast import Observations
from astroquery.sdss import SDSS
from astroquery.gaia import Gaia
from drizzlepac import tweakreg, astrodrizzle
from drizzlepac.processInput import getMdriztabPars
%config InlineBackend.figure_format = 'retina' # Improves the resolution of figures rendered in notebooks.
Gaia.MAIN_GAIA_TABLE = 'gaiadr3.gaia_source' # Change this to the desired Gaia data release.
Gaia.ROW_LIMIT = 100000 # Show all of the matched sources in the printed tables.
# Set the locations of reference files and retrieve the MDRIZTAB recommended drizzle parameters.
os.environ['CRDS_SERVER_URL'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_SERVER'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_PATH'] = './crds_cache'
os.environ['iref'] = './crds_cache/references/hst/wfc3/'
1. Download the Observations with astroquery#
For this notebook, we will download WFC3/UVIS images of NGC 6791 from program 12692 in the F606W filter acquired in Visit 01. The three FLC images have been processed by the HST WFC3 pipeline (calwf3), which includes bias subtraction, dark current and CTE correction, cosmic-ray rejection, and flat-fielding:
ibwb01xqq_flc.fits
ibwb01xrq_flc.fits
ibwb01xxq_flc.fits
MAST queries may be done using `query_criteria`, where we specify:
\(\rightarrow\) obs_id, proposal_id, and filters
MAST data products may be downloaded by using download_products, where we specify:
\(\rightarrow\) products = calibrated (FLT, FLC) or drizzled (DRZ, DRC) files
\(\rightarrow\) type = standard products (CALxxx) or advanced products (HAP-SVM)
obs_ids = ['ibwb01*']
props = ['12692']
filts = ['F606W']
obsTable = Observations.query_criteria(obs_id=obs_ids, proposal_id=props, filters=filts)
products = Observations.get_product_list(obsTable)
data_prod = ['FLC'] # ['FLC','FLT','DRC','DRZ']
data_type = ['CALWF3'] # ['CALACS','CALWF3','CALWP2','HAP-SVM']
Observations.download_products(products, productSubGroupDescription=data_prod, project=data_type, cache=True)
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibwb01xqq_flc.fits to ./mastDownload/HST/ibwb01xqq/ibwb01xqq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibwb01xrq_flc.fits to ./mastDownload/HST/ibwb01xrq/ibwb01xrq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibwb01xxq_flc.fits to ./mastDownload/HST/ibwb01xxq/ibwb01xxq_flc.fits ...
[Done]
| Local Path | Status | Message | URL |
|---|---|---|---|
| str47 | str8 | object | object |
| ./mastDownload/HST/ibwb01xqq/ibwb01xqq_flc.fits | COMPLETE | None | None |
| ./mastDownload/HST/ibwb01xrq/ibwb01xrq_flc.fits | COMPLETE | None | None |
| ./mastDownload/HST/ibwb01xxq/ibwb01xxq_flc.fits | COMPLETE | None | None |
Move to the files to the local working directory:
for flc in glob.glob('./mastDownload/HST/*/*flc.fits'):
flc_name = os.path.basename(flc)
os.rename(flc, flc_name)
if os.path.exists('mastDownload'):
shutil.rmtree('mastDownload')
1.1 Check image header data#
The cell below retrieves values for specific keywords from the first and second extensions of the image header. We see that the 1st exposure is 30 seconds and the 2nd and 3rd exposures are 360 seconds in duration. The 3rd exposure is dithered by approximately 82 arcseconds in the y-direction which is approximately the width of one WFC3 UVIS detector chip. A full list of header keywords is available in Section 2.4 of the WFC3 Data Handbook.
paths = sorted(glob.glob('*flc.fits'))
data = []
keywords_ext0 = ["ROOTNAME", "ASN_ID", "TARGNAME", "DETECTOR", "FILTER", "exptime",
"ra_targ", "dec_targ", "postarg1", "postarg2", "DATE-OBS"]
keywords_ext1 = ["orientat"]
for path in paths:
path_data = []
for keyword in keywords_ext0:
path_data.append(fits.getval(path, keyword, ext=0))
for keyword in keywords_ext1:
path_data.append(fits.getval(path, keyword, ext=1))
data.append(path_data)
keywords = keywords_ext0 + keywords_ext1
table = Table(np.array(data), names=keywords, dtype=['str', 'str', 'str', 'str', 'str', 'f8', 'f8', 'f8', 'f8', 'f8', 'str', 'f8'])
table['exptime'].format = '7.1f'
table['ra_targ'].format = table['dec_targ'].format = '7.4f'
table['postarg1'].format = table['postarg2'].format = '7.3f'
table['orientat'].format = '7.2f'
table
| ROOTNAME | ASN_ID | TARGNAME | DETECTOR | FILTER | exptime | ra_targ | dec_targ | postarg1 | postarg2 | DATE-OBS | orientat |
|---|---|---|---|---|---|---|---|---|---|---|---|
| str32 | str32 | str32 | str32 | str32 | float64 | float64 | float64 | float64 | float64 | str32 | float64 |
| ibwb01xqq | NONE | NGC-6791 | UVIS | F606W | 30.0 | 290.2248 | 37.8027 | 0.000 | 0.000 | 2011-10-20 | -56.27 |
| ibwb01xrq | NONE | NGC-6791 | UVIS | F606W | 360.0 | 290.2248 | 37.8027 | 0.000 | 0.000 | 2011-10-20 | -56.27 |
| ibwb01xxq | NONE | NGC-6791 | UVIS | F606W | 360.0 | 290.2248 | 37.8027 | 0.000 | 81.600 | 2011-10-20 | -56.25 |
1.2 Inspect the Alignment#
Check the active WCS solution in the image header. If the image is aligned to a catalog, the list the number of matches and the fit RMS in milli-arcseconds. Next, convert the fit RMS values to pixels for comparison with the alignment results performed later in this notebook using Tweakreg.
ext_0_keywords = ['DETECTOR', 'EXPTIME'] # extension 0 keywords.
ext_1_keywords = ['WCSNAME', 'NMATCHES', 'RMS_RA', 'RMS_DEC'] # extension 1 keywords.
# Define the detector plate scales in arcsec per pixel.
DETECTOR_SCALES = {
'IR': 0.1283,
'UVIS': 0.0396,
'WFC': 0.05
}
formatted_data = {}
column_data = defaultdict(list)
for fits_file in sorted(glob.glob('*flc.fits')):
column_data['filename'].append(fits_file)
header0 = fits.getheader(fits_file, 0)
header1 = fits.getheader(fits_file, 1)
for keyword in ext_0_keywords:
column_data[keyword].append(header0[keyword])
for keyword in ext_1_keywords:
if 'RMS' in keyword:
value = np.around(header1[keyword], decimals=1)
else:
value = header1[keyword]
column_data[keyword].append(value)
for keyword in ['RMS_RA', 'RMS_DEC']:
rms_value = header1[keyword] / 1000 / DETECTOR_SCALES[header0['DETECTOR']]
column_data[f'{keyword}_pix'].append(np.round(rms_value, decimals=2))
wcstable = Table(column_data)
wcstable
| filename | DETECTOR | EXPTIME | WCSNAME | NMATCHES | RMS_RA | RMS_DEC | RMS_RA_pix | RMS_DEC_pix |
|---|---|---|---|---|---|---|---|---|
| str18 | str4 | float64 | str30 | int64 | float64 | float64 | float64 | float64 |
| ibwb01xqq_flc.fits | UVIS | 30.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 435 | 2.4 | 2.3 | 0.06 | 0.06 |
| ibwb01xrq_flc.fits | UVIS | 360.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 456 | 3.6 | 3.2 | 0.09 | 0.08 |
| ibwb01xxq_flc.fits | UVIS | 360.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 429 | 3.9 | 3.8 | 0.1 | 0.1 |
2. Query the Reference Catalogs#
Now that we have the FITS images, we will download the reference catalogs from both SDSS and Gaia using astroquery.
2.1 Identify Coordinates#
We will first create a SkyCoord Object to point astroquery to where we are looking on the sky. Since our example uses data from NGC 6791, we will use the ra_targ and dec_targ keywords from the first image to get the coordinates of the object.
RA = table['ra_targ'][0]
Dec = table['dec_targ'][0]
2.2 SDSS Query#
We now give the RA and Dec values to an astropy SkyCoord object, which we will pass to the SDSS query. Additionally, we use an astropy Quantity object to create a radius for the SDSS query in physical units. For reference, the UVIS detector field of view is approximately 2.7’\(\times\)2.7’ and a y-dither of 82 arcseconds covers a total area on the sky of approximately 2.7’\(\times\)4.1’. For SDSS the maximum search radius is 3 arcminutes, which covers the majority of our field of view in this example. In the Gaia query, we set the radius to 5 arcminutes to fully cover the area of our observations.
coord = SkyCoord(ra=RA, dec=Dec, unit=(u.deg, u.deg))
Next, we perform the query via the SDSS.query_region method of astroquery.sdss. The spectro=False keyword argument means we want to exclude spectroscopic objects, as we are looking for objects to match with an image. In the fields parameter, we specify a list of fields we want returned by the query. In this case we only need the position, as well as a g-band magnitude if we want to remove very dim and/or bright objects from the catalog, as those are likely measured poorly. Details on selecting objects by magnitude may be found in the ‘Gaia_alignment’ notebook. Many other fields are available in the SDSS.query_region function, which are documented on this webpage.
radius = Quantity(3., u.arcmin)
sdss_query = SDSS.query_region(coord, radius=radius, spectro=False, fields=['ra', 'dec', 'g'])
sdss_query
| ra | dec | g |
|---|---|---|
| float64 | float64 | float64 |
| 290.175257483306 | 37.7757023742629 | 19.10387 |
| 290.175260270816 | 37.8293581828758 | -9999.0 |
| 290.175268441296 | 37.7757076932531 | 18.98614 |
| 290.175307121632 | 37.7548760313867 | 19.49425 |
| 290.175317556537 | 37.8293308028686 | 18.26527 |
| 290.175318784872 | 37.8293349727083 | -9999.0 |
| 290.175320686698 | 37.7548746619173 | 19.7736 |
| 290.175328125755 | 37.7625936217729 | 19.44313 |
| 290.175333152015 | 37.8293270812477 | 18.38934 |
| ... | ... | ... |
| 290.274703632708 | 37.7877877835155 | 18.8502 |
| 290.274704603188 | 37.7891141064558 | 18.63407 |
| 290.274721361389 | 37.7877737666928 | 19.01597 |
| 290.27472930088 | 37.7890657093451 | 18.74797 |
| 290.274794783948 | 37.7993797493131 | 16.67577 |
| 290.274800891914 | 37.7993938284574 | 16.11324 |
| 290.274807588715 | 37.799392577291 | 16.08992 |
| 290.274815152976 | 37.7993736308255 | 16.13595 |
| 290.27482391543 | 37.7993757145458 | 16.16263 |
| 290.274839705192 | 37.8423495653678 | 20.26493 |
We then need to save the catalog to use with TweakReg. As the returned value of the query is an astropy.Table, saving the file is straightforward:
sdss_query.write('sdss.cat', format='ascii.commented_header', overwrite=True)
2.3 Gaia Query#
Similarly to SDSS, we can query Gaia catalogs for our target via astroquery.gaia. This may be preferable in many cases because the spaced-based astrometry from Gaia is generally very accurate and precise. We can use the same coord search parameters from our earlier SDSS query, along with a slightly larger radius that encompasses the full field of view of these observations.
radius = Quantity(5., u.arcmin)
gaia_query = Gaia.query_object_async(coordinate=coord, radius=radius)
gaia_query
INFO: Query finished. [astroquery.utils.tap.core]
| solution_id | DESIGNATION | SOURCE_ID | random_index | ref_epoch | ra | ra_error | dec | dec_error | parallax | parallax_error | parallax_over_error | pm | pmra | pmra_error | pmdec | pmdec_error | ra_dec_corr | ra_parallax_corr | ra_pmra_corr | ra_pmdec_corr | dec_parallax_corr | dec_pmra_corr | dec_pmdec_corr | parallax_pmra_corr | parallax_pmdec_corr | pmra_pmdec_corr | astrometric_n_obs_al | astrometric_n_obs_ac | astrometric_n_good_obs_al | astrometric_n_bad_obs_al | astrometric_gof_al | astrometric_chi2_al | astrometric_excess_noise | astrometric_excess_noise_sig | astrometric_params_solved | astrometric_primary_flag | nu_eff_used_in_astrometry | pseudocolour | pseudocolour_error | ra_pseudocolour_corr | dec_pseudocolour_corr | parallax_pseudocolour_corr | pmra_pseudocolour_corr | pmdec_pseudocolour_corr | astrometric_matched_transits | visibility_periods_used | astrometric_sigma5d_max | matched_transits | new_matched_transits | matched_transits_removed | ipd_gof_harmonic_amplitude | ipd_gof_harmonic_phase | ipd_frac_multi_peak | ipd_frac_odd_win | ruwe | scan_direction_strength_k1 | scan_direction_strength_k2 | scan_direction_strength_k3 | scan_direction_strength_k4 | scan_direction_mean_k1 | scan_direction_mean_k2 | scan_direction_mean_k3 | scan_direction_mean_k4 | duplicated_source | phot_g_n_obs | phot_g_mean_flux | phot_g_mean_flux_error | phot_g_mean_flux_over_error | phot_g_mean_mag | phot_bp_n_obs | phot_bp_mean_flux | phot_bp_mean_flux_error | phot_bp_mean_flux_over_error | phot_bp_mean_mag | phot_rp_n_obs | phot_rp_mean_flux | phot_rp_mean_flux_error | phot_rp_mean_flux_over_error | phot_rp_mean_mag | phot_bp_rp_excess_factor | phot_bp_n_contaminated_transits | phot_bp_n_blended_transits | phot_rp_n_contaminated_transits | phot_rp_n_blended_transits | phot_proc_mode | bp_rp | bp_g | g_rp | radial_velocity | radial_velocity_error | rv_method_used | rv_nb_transits | rv_nb_deblended_transits | rv_visibility_periods_used | rv_expected_sig_to_noise | rv_renormalised_gof | rv_chisq_pvalue | rv_time_duration | rv_amplitude_robust | rv_template_teff | rv_template_logg | rv_template_fe_h | rv_atm_param_origin | vbroad | vbroad_error | vbroad_nb_transits | grvs_mag | grvs_mag_error | grvs_mag_nb_transits | rvs_spec_sig_to_noise | phot_variable_flag | l | b | ecl_lon | ecl_lat | in_qso_candidates | in_galaxy_candidates | non_single_star | has_xp_continuous | has_xp_sampled | has_rvs | has_epoch_photometry | has_epoch_rv | has_mcmc_gspphot | has_mcmc_msc | in_andromeda_survey | classprob_dsc_combmod_quasar | classprob_dsc_combmod_galaxy | classprob_dsc_combmod_star | teff_gspphot | teff_gspphot_lower | teff_gspphot_upper | logg_gspphot | logg_gspphot_lower | logg_gspphot_upper | mh_gspphot | mh_gspphot_lower | mh_gspphot_upper | distance_gspphot | distance_gspphot_lower | distance_gspphot_upper | azero_gspphot | azero_gspphot_lower | azero_gspphot_upper | ag_gspphot | ag_gspphot_lower | ag_gspphot_upper | ebpminrp_gspphot | ebpminrp_gspphot_lower | ebpminrp_gspphot_upper | libname_gspphot | dist |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| yr | deg | mas | deg | mas | mas | mas | mas / yr | mas / yr | mas / yr | mas / yr | mas / yr | mas | 1 / um | 1 / um | 1 / um | mas | deg | deg | deg | deg | deg | electron / s | electron / s | mag | electron / s | electron / s | mag | electron / s | electron / s | mag | mag | mag | mag | km / s | km / s | d | km / s | K | log(cm.s**-2) | dex | km / s | km / s | mag | mag | deg | deg | deg | deg | K | K | K | log(cm.s**-2) | log(cm.s**-2) | log(cm.s**-2) | dex | dex | dex | pc | pc | pc | mag | mag | mag | mag | mag | mag | mag | mag | mag | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| int64 | object | int64 | int64 | float64 | float64 | float32 | float64 | float32 | float64 | float32 | float32 | float32 | float64 | float32 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | int16 | bool | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | float32 | int16 | int16 | int16 | float32 | float32 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | bool | int16 | float64 | float32 | float32 | float32 | int16 | float64 | float32 | float32 | float32 | int16 | float64 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | float32 | float32 | int16 | float32 | float32 | int16 | float32 | object | float64 | float64 | float64 | float64 | bool | bool | int16 | bool | bool | bool | bool | bool | bool | bool | bool | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | object | float64 |
| 1636148068921376768 | Gaia DR3 2051293319932402944 | 2051293319932402944 | 1710414812 | 2016.0 | 290.2248611356283 | 0.12343551 | 37.80192950361359 | 0.15534069 | 1.2409154633064086 | 0.15976872 | 7.7669487 | 6.0644784 | -0.6226543407618474 | 0.15474372 | 6.0324290792879545 | 0.18924683 | -0.033001468 | 0.16417459 | -0.027278472 | -0.020170886 | 0.07942047 | -0.046242356 | -0.19703287 | -0.07685185 | 0.0051713893 | 0.050814763 | 394 | 0 | 389 | 5 | 0.407836 | 413.48303 | 0.0 | 2.0511602e-15 | 31 | False | 1.3016587 | -- | -- | -- | -- | -- | -- | -- | 45 | 26 | 0.26493552 | 49 | 22 | 0 | 0.01895016 | 14.319122 | 0 | 0 | 1.0138804 | 0.17569481 | 0.20240329 | 0.10288032 | 0.089386284 | -95.13182 | 77.09379 | 2.6807811 | -30.180996 | False | 385 | 492.16788454743386 | 0.9903239 | 496.97668 | 18.957083 | 39 | 106.26453488321661 | 5.3855076 | 19.731573 | 20.272572 | 39 | 549.1994295106254 | 7.0852404 | 77.51317 | 17.898571 | 1.3317894 | 0 | 2 | 0 | 2 | 0 | 2.3740005 | 1.3154888 | 1.0585117 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98761891051129 | 10.914199739506339 | 302.04196574777654 | 59.01190915812673 | False | False | 0 | False | False | False | False | False | True | False | False | 1.4003378e-13 | 5.356044e-13 | 0.99996775 | 3653.592 | 3591.5752 | 3744.3992 | 4.8552 | 4.4992 | 4.9965 | -0.4967 | -0.7199 | -0.1315 | 653.0443 | 491.9481 | 1283.5112 | 0.7812 | 0.6499 | 0.9202 | 0.5305 | 0.44 | 0.6266 | 0.3102 | 0.2581 | 0.3649 | MARCS | 0.0007449136479797354 |
| 1636148068921376768 | Gaia DR3 2051293324218263808 | 2051293324218263808 | 214974978 | 2016.0 | 290.22327972754084 | 1.0491472 | 37.80283944795903 | 1.4068799 | -0.6069077460064267 | 1.4172788 | -0.42822045 | 3.8347042 | -0.49694994391852737 | 1.433404 | -3.802367186693702 | 2.183717 | -0.32559526 | 0.44083008 | 0.040013984 | -0.36232764 | -0.18673699 | -0.29855755 | 0.5145219 | 0.16053891 | -0.3253903 | -0.32209665 | 133 | 0 | 132 | 1 | 2.3624072 | 187.53317 | 3.4946198 | 1.7356544 | 95 | False | -- | 1.2583821 | 0.3532982 | -0.10796609 | -0.06725511 | -0.030691369 | -0.22952616 | -0.12757729 | 16 | 14 | 3.2688706 | 18 | 6 | 1 | 0.061227437 | 43.226418 | 5 | 17 | 1.1496774 | 0.23548113 | 0.3243092 | 0.35748923 | 0.4221966 | -95.12672 | 74.99657 | -8.526287 | -16.614477 | False | 132 | 88.7375766294378 | 1.1992922 | 73.99162 | 20.817099 | 0 | -- | -- | -- | -- | 0 | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.9878943903652 | 10.915721621139165 | 302.0400990046131 | 59.01311977354264 | False | False | 0 | False | False | False | False | False | False | False | False | 7.759076e-08 | 3.7895185e-09 | 0.9999943 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.0012108975043825237 | |
| 1636148068921376768 | Gaia DR3 2051293324222972288 | 2051293324222972288 | 164456380 | 2016.0 | 290.22261994880597 | 0.032841645 | 37.80270752474709 | 0.039459 | 0.3053094896365258 | 0.041127697 | 7.423452 | 2.5961652 | -0.7012623275324616 | 0.041221105 | -2.4996610433857405 | 0.047550496 | -0.047555298 | 0.16546503 | 0.021742897 | -0.077867456 | 0.066915035 | -0.08765522 | -0.26260835 | -0.059347805 | -0.087691456 | -0.0013310307 | 383 | 0 | 381 | 2 | -2.1703348 | 333.38327 | 0.0 | 0.0 | 31 | False | 1.4413656 | -- | -- | -- | -- | -- | -- | -- | 44 | 25 | 0.06704042 | 48 | 21 | 0 | 0.015037518 | 12.04822 | 0 | 0 | 0.9210467 | 0.13890518 | 0.18901813 | 0.10900513 | 0.0641752 | -88.36165 | 72.302956 | 5.923747 | -33.787945 | False | 381 | 4179.254659077206 | 2.2517574 | 1855.9968 | 16.63462 | 43 | 1717.7830387470858 | 6.2028065 | 276.93643 | 17.251122 | 39 | 3494.1204079650242 | 10.008291 | 349.1226 | 15.889551 | 1.2470893 | 0 | 25 | 0 | 23 | 0 | 1.3615704 | 0.61650085 | 0.7450695 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98754100993882 | 10.916132432650413 | 302.0390547900942 | 59.01313187059839 | False | False | 0 | True | False | False | False | False | True | True | False | 1.0228448e-13 | 9.3310026e-11 | 0.9999741 | 4770.3447 | 4749.6045 | 4790.818 | 4.1776 | 4.1596 | 4.1902 | 0.3546 | 0.3268 | 0.3805 | 2185.9316 | 2147.3625 | 2239.1926 | 0.3511 | 0.3259 | 0.3735 | 0.2701 | 0.2505 | 0.2875 | 0.1406 | 0.1303 | 0.1496 | PHOENIX | 0.001721026645501702 |
| 1636148068921376768 | Gaia DR3 2051293324222972416 | 2051293324222972416 | 1599905588 | 2016.0 | 290.2248309701555 | 0.052941296 | 37.80058611405895 | 0.064896055 | 0.21837043824644525 | 0.067057766 | 3.2564526 | 2.3448853 | -0.5827200539782851 | 0.066188715 | -2.2713266001611636 | 0.0787358 | -0.076830104 | 0.14136231 | -0.025055349 | -0.07297541 | 0.056257915 | -0.07904705 | -0.24559797 | -0.07446872 | -0.087429665 | -0.03352803 | 395 | 0 | 392 | 3 | -0.037620842 | 401.51434 | 0.0 | 0.0 | 31 | False | 1.4891585 | -- | -- | -- | -- | -- | -- | -- | 45 | 25 | 0.110738866 | 49 | 22 | 0 | 0.0153436065 | 15.8542 | 0 | 0 | 0.99778724 | 0.15293275 | 0.21477312 | 0.12928022 | 0.08397196 | -94.649994 | 70.08022 | 6.0968003 | -29.722376 | False | 389 | 1751.0366076397277 | 1.4950225 | 1171.2443 | 17.579128 | 46 | 817.0065234341104 | 6.4622293 | 126.42797 | 18.057978 | 44 | 1347.155230450357 | 8.58573 | 156.90633 | 16.92435 | 1.2359318 | 0 | 32 | 0 | 29 | 0 | 1.1336269 | 0.4788494 | 0.6547775 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98637845348246 | 10.913632562752337 | 302.0412242635996 | 59.01062093567249 | False | False | 0 | True | False | False | False | False | True | True | False | 6.110852e-11 | 5.145215e-13 | 0.9999132 | 4831.901 | 4812.959 | 4859.14 | 4.661 | 4.6412 | 4.6768 | -0.4447 | -0.5218 | -0.3786 | 1595.5184 | 1545.6223 | 1641.4886 | 0.0087 | 0.0021 | 0.021 | 0.0069 | 0.0017 | 0.0166 | 0.0036 | 0.0009 | 0.0088 | PHOENIX | 0.0020867793124382814 |
| 1636148068921376768 | Gaia DR3 2051293319934791936 | 2051293319934791936 | 127740795 | 2016.0 | 290.2225602638809 | 0.098567784 | 37.80397847004457 | 0.12238157 | 0.18608923760796978 | 0.12619987 | 1.4745597 | 2.3556678 | -0.20591766034379622 | 0.12384657 | -2.346650638048337 | 0.14836667 | -0.0034372064 | 0.17569284 | -0.028029243 | -0.023688797 | 0.0831415 | -0.04713238 | -0.21456409 | -0.08000817 | -0.019006517 | 0.07798844 | 386 | 0 | 383 | 3 | 0.34466058 | 403.29874 | 0.0 | 0.0 | 31 | False | 1.4745288 | -- | -- | -- | -- | -- | -- | -- | 44 | 26 | 0.20897423 | 48 | 21 | 0 | 0.02525984 | 16.266506 | 0 | 0 | 1.011676 | 0.16451268 | 0.180577 | 0.079406776 | 0.08114059 | -89.370926 | 80.65579 | 0.7836158 | -30.156015 | False | 377 | 665.1667185914472 | 1.124127 | 591.71844 | 18.630041 | 40 | 296.01604570644196 | 5.8455243 | 50.63978 | 19.160254 | 41 | 517.3148686523093 | 6.1276155 | 84.42352 | 17.963509 | 1.2227474 | 0 | 0 | 0 | 0 | 0 | 1.1967449 | 0.5302124 | 0.6665325 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98868349525281 | 10.916731652994313 | 302.03962583571473 | 59.014369244472704 | False | False | 0 | False | False | False | False | False | True | False | False | 1.0210282e-13 | 5.259736e-13 | 0.99999475 | 4590.2783 | 4575.766 | 4603.0244 | 4.8255 | 4.7981 | 4.858 | -1.2317 | -1.4162 | -1.1009 | 1664.2876 | 1569.1196 | 1730.915 | 0.0102 | 0.0026 | 0.0257 | 0.008 | 0.002 | 0.02 | 0.0043 | 0.0011 | 0.0107 | PHOENIX | 0.0021977590777958536 |
| 1636148068921376768 | Gaia DR3 2051293324216543232 | 2051293324216543232 | 496974628 | 2016.0 | 290.22681959687196 | 0.5800434 | 37.80112772770517 | 0.8518114 | 0.9866119688573637 | 0.8487393 | 1.162444 | 4.5651155 | -0.44243766262405015 | 1.042491 | -4.543625170019005 | 1.2715639 | -0.03034182 | -0.13035429 | 0.09905615 | 0.024711896 | -0.035745695 | -0.13054979 | 0.34666777 | -0.069062054 | -0.28526154 | -0.1078611 | 189 | 0 | 189 | 0 | -0.84486425 | 177.7449 | 0.0 | 1.4856265e-15 | 95 | False | -- | 1.5521348 | 0.21582161 | -0.0062133768 | 0.2804249 | 0.035700142 | -0.42101106 | 0.17818592 | 22 | 15 | 1.8150634 | 25 | 8 | 0 | 0.023252329 | 30.515776 | 0 | 0 | 0.954371 | 0.20877054 | 0.28841907 | 0.1927084 | 0.43569437 | -76.9166 | 84.362335 | -3.7503886 | -19.752798 | False | 189 | 100.17345952653844 | 0.81711596 | 122.59393 | 20.685486 | 16 | 34.777743067191494 | 8.345614 | 4.1671877 | 21.485289 | 19 | 89.51229139692614 | 5.5230875 | 16.20693 | 19.868189 | 1.2407482 | 0 | 0 | 0 | 0 | 0 | 1.6170998 | 0.7998028 | 0.817297 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98757543345182 | 10.912457430119012 | 302.0444461542871 | 59.01072318960171 | False | False | 0 | False | False | False | False | False | False | False | False | 1.5372552e-09 | 1.1275478e-09 | 0.99999505 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.002222460820739603 | |
| 1636148068921376768 | Gaia DR3 2051293324222974464 | 2051293324222974464 | 1798436105 | 2016.0 | 290.2275249308142 | 0.24079925 | 37.803472775139035 | 0.3074705 | 0.44506652441996963 | 0.3134235 | 1.4200164 | 1.903844 | -0.7741499247735619 | 0.3071843 | -1.7393429825276292 | 0.37454808 | 0.020487515 | 0.17073557 | -0.036337543 | 0.041650914 | 0.12431544 | -0.0061035734 | -0.18864702 | -0.06549308 | 0.060529068 | 0.123350315 | 382 | 0 | 380 | 2 | 0.5945536 | 412.03198 | 0.9015461 | 0.9578774 | 95 | False | -- | 1.3147619 | 0.081987195 | -0.016766502 | 0.118190974 | 0.037860535 | -0.0898746 | -0.10861043 | 44 | 25 | 0.52942884 | 48 | 21 | 0 | 0.027762208 | 20.245789 | 0 | 4 | 1.02092 | 0.18400808 | 0.1789637 | 0.073698275 | 0.10250232 | -90.673706 | 85.22672 | -9.854181 | -30.444962 | False | 380 | 207.20873617672416 | 0.7419642 | 279.2705 | 19.896347 | 9 | 91.39950015947561 | 7.5014834 | 12.18419 | 20.436182 | 9 | 224.20190021064204 | 11.064253 | 20.263628 | 18.871298 | 1.5231086 | 0 | 0 | 0 | 0 | 0 | 1.5648842 | 0.539835 | 1.0250492 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98997089407423 | 10.912983986333725 | 302.04670590522085 | 59.01283416072846 | False | False | 0 | False | False | False | False | False | False | False | False | 1.8206678e-09 | 2.9034798e-08 | 0.99998796 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.002298594469020544 | |
| 1636148068921376768 | Gaia DR3 2051293324216539648 | 2051293324216539648 | 1649457609 | 2016.0 | 290.2242320409821 | 0.5271274 | 37.80039510739463 | 0.76655334 | 1.3632756584241434 | 0.6728546 | 2.0261073 | 2.2253373 | -0.9708645882906146 | 0.6578662 | -2.0023856302519407 | 0.98862535 | -0.33465353 | 0.08556003 | -0.2735162 | 0.2602703 | 0.161614 | 0.21899539 | -0.26542807 | 0.033500012 | 0.1458301 | -0.23663591 | 243 | 0 | 243 | 0 | 1.2024069 | 273.66382 | 0.0 | 0.0 | 95 | False | -- | 1.6818463 | 0.19114311 | -0.26075235 | 0.11348202 | -0.08333635 | -0.06321393 | -0.15212661 | 28 | 18 | 1.4304484 | 32 | 14 | 1 | 0.0707902 | 34.375954 | 0 | 15 | 1.0543019 | 0.24560685 | 0.4438982 | 0.22968337 | 0.165783 | -110.25776 | 67.43046 | 5.994361 | -23.29821 | False | 244 | 109.8552156027335 | 0.8030061 | 136.80496 | 20.585316 | 0 | -- | -- | -- | -- | 0 | -- | -- | -- | -- | -- | -- | -- | -- | -- | 2 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98599242480627 | 10.91397427439119 | 302.0402394379523 | 59.01056325889003 | False | False | 0 | False | False | False | False | False | False | False | False | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.0023210548670645637 | |
| 1636148068921376768 | Gaia DR3 2051293324225336064 | 2051293324225336064 | 398090960 | 2016.0 | 290.22302939666463 | 0.47650018 | 37.8003404115494 | 0.57163584 | 1.0465539617532613 | 0.60573983 | 1.7277284 | 0.3426481 | 0.016525679687419553 | 0.6219764 | -0.34224935447443744 | 0.7205138 | -0.11358616 | 0.1529165 | 0.07183364 | -0.104466826 | 0.054135017 | -0.07860708 | -0.1758327 | 0.10776901 | -0.16631071 | -0.16472225 | 373 | 0 | 365 | 8 | 86.80703 | 12487.478 | 6.598426 | 424.3264 | 95 | False | -- | 1.2976544 | 0.14454299 | -0.057665557 | 0.052674405 | 0.033202734 | 0.010857275 | -0.07922697 | 43 | 25 | 1.0361372 | 50 | 50 | 0 | 0.19814193 | 21.407808 | 63 | 0 | 5.6149945 | 0.1693135 | 0.21580335 | 0.10183408 | 0.09376588 | -92.04377 | 70.39044 | 0.82470447 | -23.783632 | False | 298 | 836.206629484886 | 5.043918 | 165.78514 | 18.381582 | 43 | 492.85805036589596 | 5.0838623 | 96.945595 | 18.606737 | 41 | 916.8429900655037 | 6.7629867 | 135.56776 | 17.342157 | 1.6858286 | 0 | 2 | 0 | 2 | 0 | 1.2645798 | 0.22515488 | 1.0394249 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.98551833187275 | 10.914804515213548 | 302.0384324817961 | 59.01076433315906 | False | False | 0 | False | False | False | False | False | False | False | False | 9.514203e-11 | 1.056036e-05 | 0.99998164 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.0027187891427339723 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1636148068921376768 | Gaia DR3 2051294870411535616 | 2051294870411535616 | 430955701 | 2016.0 | 290.1710542799103 | 0.089351624 | 37.874210138824154 | 0.10865699 | 0.3694586266163912 | 0.113834515 | 3.2455766 | 9.114665 | -6.31871143886455 | 0.11210035 | -6.568942750936787 | 0.13436429 | -0.007841255 | 0.11561305 | -0.01169015 | -0.03635325 | 0.11398082 | -0.05428327 | -0.21885028 | -0.10007106 | -0.08220788 | 0.061571144 | 399 | 0 | 392 | 7 | 0.13260055 | 406.64862 | 0.22923899 | 0.41862753 | 31 | False | 1.4996165 | -- | -- | -- | -- | -- | -- | -- | 46 | 27 | 0.18900031 | 49 | 22 | 0 | 0.011167029 | 1.7622564 | 0 | 0 | 1.0039074 | 0.26786533 | 0.10760525 | 0.10404573 | 0.08249484 | -87.45818 | 75.753624 | -15.217908 | -38.780235 | False | 384 | 779.8054003243237 | 1.0609461 | 735.00946 | 18.457401 | 46 | 362.8243922628937 | 6.342421 | 57.20598 | 18.9393 | 45 | 563.964440036763 | 5.256008 | 107.299 | 17.869766 | 1.1884873 | 0 | 0 | 0 | 0 | 0 | 1.0695343 | 0.48189926 | 0.58763504 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 70.03485933670954 | 10.984050062538527 | 301.9998719659017 | 59.09291303817852 | False | False | 0 | False | False | False | False | False | True | False | False | 3.5958955e-11 | 5.3623333e-13 | 0.9999949 | 4889.5947 | 4878.3716 | 4901.4575 | 4.8392 | 4.8272 | 4.8515 | -1.6225 | -1.7671 | -1.5237 | 1828.6827 | 1778.3691 | 1860.6678 | 0.0045 | 0.0011 | 0.0108 | 0.0036 | 0.0009 | 0.0086 | 0.002 | 0.0005 | 0.0047 | MARCS | 0.0831808340919906 |
| 1636148068921376768 | Gaia DR3 2051288234680628736 | 2051288234680628736 | 275359989 | 2016.0 | 290.1228125225418 | 0.12832773 | 37.7819163631625 | 0.16770653 | 0.45711970927190115 | 0.16442272 | 2.7801495 | 15.87947 | -6.491827713462847 | 0.16428103 | -14.491851015938131 | 0.21764661 | 0.002764468 | 0.12523003 | -0.10028199 | 0.050657623 | 0.06666436 | 0.005579139 | -0.37272593 | -0.118829906 | 0.0050995317 | 0.1139823 | 409 | 0 | 409 | 0 | -1.3663982 | 383.07782 | 0.0 | 1.9997444e-15 | 31 | False | 1.3034092 | -- | -- | -- | -- | -- | -- | -- | 47 | 27 | 0.3113583 | 48 | 23 | 0 | 0.010293381 | 2.4593005 | 0 | 0 | 0.95150584 | 0.17092009 | 0.1387889 | 0.05828934 | 0.13473459 | -116.178055 | 83.69419 | 2.6145937 | -36.532135 | False | 407 | 450.44352750937213 | 0.8725948 | 516.2116 | 19.053267 | 45 | 103.68771152397954 | 5.136524 | 20.186357 | 20.299225 | 45 | 502.45009105705196 | 3.6524675 | 137.56456 | 17.995163 | 1.3456466 | 0 | 1 | 0 | 0 | 0 | 2.304062 | 1.2459583 | 1.0581036 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.9333371194973 | 10.977945740748442 | 301.88063476010194 | 59.014107067896354 | False | False | 0 | False | False | False | False | False | False | False | False | 3.8000676e-13 | 7.05338e-13 | 0.9998841 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08322243598195858 | |
| 1636148068921376768 | Gaia DR3 2051294870411542272 | 2051294870411542272 | 1651247147 | 2016.0 | 290.1750101665401 | 0.06694008 | 37.87602842402639 | 0.092485614 | 0.20603759179502212 | 0.08579865 | 2.4014082 | 2.2515461 | -0.3932337230718288 | 0.08297268 | -2.216941094221096 | 0.118552975 | -0.06297449 | 0.14318395 | -0.033632368 | 0.03577277 | -0.034115147 | 0.00034698079 | -0.4515389 | -0.10937831 | 0.07231181 | 0.009728736 | 377 | 0 | 374 | 3 | 0.06470177 | 384.27502 | 0.0 | 0.0 | 95 | False | -- | 1.4371054 | 0.023171378 | -0.16051838 | 0.025070293 | -0.010794556 | -0.07607864 | 0.028111283 | 43 | 26 | 0.17073497 | 48 | 22 | 1 | 0.01102003 | 161.05574 | 0 | 15 | 1.0014795 | 0.2962618 | 0.18218416 | 0.14427209 | 0.11568721 | -84.172356 | 75.77043 | -18.850288 | 42.884228 | False | 374 | 1241.344401787872 | 1.2767242 | 972.2886 | 17.952637 | 0 | -- | -- | -- | -- | 0 | -- | -- | -- | -- | -- | -- | -- | -- | -- | 2 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 70.0379165005182 | 10.982037682530848 | 302.00667588191897 | 59.093831343557795 | False | False | 0 | False | False | False | False | False | False | False | False | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08322879189943044 | |
| 1636148068921376768 | Gaia DR3 2051295381516623744 | 2051295381516623744 | 1667872848 | 2016.0 | 290.2882629014902 | 0.14793906 | 37.86913015536792 | 0.21439508 | 0.6608720254317446 | 0.1987435 | 3.3252509 | 8.816373 | -2.6884990926127106 | 0.19940175 | -8.39645178415166 | 0.26135257 | -0.12174883 | 0.10869827 | 0.06210663 | 0.0064050634 | 0.07006799 | -0.034789 | -0.36298344 | 0.027279017 | -0.00647773 | -0.083680145 | 339 | 0 | 338 | 1 | 0.5362549 | 369.1253 | 0.41999847 | 0.5260669 | 95 | False | -- | 1.3389693 | 0.05077461 | -0.087569006 | 0.18131016 | -0.04555531 | -0.13339908 | -0.11010846 | 39 | 23 | 0.3739153 | 42 | 20 | 0 | 0.029168483 | 178.31316 | 0 | 0 | 1.019872 | 0.2148467 | 0.20623815 | 0.12674522 | 0.11732818 | -75.89927 | 79.33952 | -25.199 | -39.338097 | False | 346 | 420.7538148013615 | 1.0356286 | 406.27866 | 19.127296 | 39 | 140.64232775208788 | 7.4752154 | 18.814486 | 19.968252 | 39 | 478.4602734701604 | 9.109383 | 52.523895 | 18.04828 | 1.4714129 | 0 | 22 | 0 | 22 | 0 | 1.9199715 | 0.84095573 | 1.0790157 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 70.0714830724915 | 10.89864899377486 | 302.1707078793191 | 59.063253212461184 | False | False | 0 | False | False | False | False | False | False | False | False | 8.597156e-11 | 5.9048273e-09 | 0.9999822 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08324003047068324 | |
| 1636148068921376768 | Gaia DR3 2051288406479389696 | 2051288406479389696 | 856711248 | 2016.0 | 290.1194354085638 | 0.1685125 | 37.80348115009846 | 0.21949896 | 0.24010742276483804 | 0.20961256 | 1.1454821 | 6.186268 | -2.2508290547655534 | 0.21421075 | -5.762263219114228 | 0.27950856 | -0.04189369 | 0.055101015 | -0.16017503 | 0.114143774 | 0.15277705 | 0.08570732 | -0.41045648 | -0.013016244 | -0.025973871 | 0.046091143 | 369 | 0 | 369 | 0 | -0.34031537 | 371.16574 | 0.0 | 4.2135146e-15 | 31 | False | 1.4944085 | -- | -- | -- | -- | -- | -- | -- | 42 | 25 | 0.40078756 | 43 | 21 | 0 | 0.009520529 | 19.020708 | 0 | 0 | 0.9865019 | 0.20752354 | 0.112608805 | 0.099987626 | 0.1787911 | -113.13249 | 83.67482 | 12.821432 | -33.98683 | False | 367 | 332.1875262158305 | 0.86817735 | 382.62634 | 19.38391 | 38 | 136.82832781506718 | 7.0689607 | 19.356216 | 19.998102 | 40 | 250.45730102656773 | 5.909497 | 42.38217 | 18.75106 | 1.1658645 | 0 | 0 | 0 | 0 | 0 | 1.2470417 | 0.61419296 | 0.63284874 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.95190401428546 | 10.989775662227835 | 301.88677645351646 | 59.035605221497256 | False | False | 0 | False | False | False | False | False | False | False | False | 5.805757e-10 | 2.1388297e-10 | 0.9999928 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.0832529718271768 | |
| 1636148068921376768 | Gaia DR3 2051107128798566528 | 2051107128798566528 | 53785428 | 2016.0 | 290.33007288300433 | 0.5063185 | 37.799005196546766 | 0.60861003 | 2.59582303334833 | 0.72787225 | 3.5663168 | 15.07725 | -9.90581987549699 | 0.8522273 | -11.36653796175356 | 0.78330284 | -0.037682395 | -0.031865645 | -0.0061560404 | -0.0959621 | 0.24388646 | -0.024079807 | -0.085402675 | 0.35197842 | -0.08016355 | -0.08673279 | 257 | 0 | 257 | 0 | 2.9779897 | 345.6605 | 2.6002293 | 2.6564052 | 95 | False | -- | 1.3103058 | 0.1573422 | -0.10415734 | 0.033458136 | -0.09948709 | -0.2720693 | 0.04197427 | 31 | 22 | 1.2133433 | 33 | 13 | 1 | 0.020845002 | 138.17416 | 0 | 0 | 1.1344311 | 0.18316215 | 0.09937623 | 0.23592351 | 0.23159805 | -121.595795 | 82.81833 | -15.507539 | -25.80699 | False | 253 | 128.70082874164865 | 0.86452293 | 148.86919 | 20.413414 | 6 | 39.717968158819254 | 8.353689 | 4.7545424 | 21.341074 | 10 | 145.18349404606658 | 15.55031 | 9.336373 | 19.343102 | 1.4366765 | 0 | 0 | 0 | 0 | 0 | 1.9979725 | 0.92766 | 1.0703125 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 70.02207192846973 | 10.83820692421412 | 302.19596551414276 | 58.98684034920937 | False | False | 0 | False | False | False | False | False | False | False | False | 2.8843332e-12 | 1.4122469e-09 | 0.99999326 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08326359931304438 | |
| 1636148068921376768 | Gaia DR3 2051287964103728384 | 2051287964103728384 | 1638894158 | 2016.0 | 290.14278652187744 | 0.03494116 | 37.75037525259858 | 0.04241879 | 0.18366243526469542 | 0.043460846 | 4.2259283 | 2.248427 | -0.4281612111184363 | 0.04497176 | -2.2072837830215937 | 0.050941683 | 0.0351151 | 0.055712797 | -0.14853671 | -0.051824972 | 0.13105968 | -0.06969542 | -0.3188419 | -0.1026289 | -0.11638979 | 0.09372311 | 428 | 0 | 426 | 2 | -0.6057206 | 420.23416 | 0.0 | 0.0 | 31 | False | 1.5003259 | -- | -- | -- | -- | -- | -- | -- | 49 | 28 | 0.07342281 | 51 | 24 | 0 | 0.03158952 | 8.174233 | 0 | 0 | 0.97841215 | 0.12762302 | 0.13048583 | 0.06985256 | 0.09125381 | -109.3513 | 84.420715 | -1.0416744 | -31.609655 | False | 426 | 3374.6423730218935 | 1.91779 | 1759.6516 | 16.866798 | 48 | 1618.891181717306 | 6.9249954 | 233.77505 | 17.315498 | 49 | 2463.8001524600472 | 6.911771 | 356.4644 | 16.268883 | 1.2098145 | 0 | 2 | 0 | 1 | 0 | 1.0466156 | 0.44869995 | 0.59791565 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.9114824714278 | 10.949949126990399 | 301.8939100069244 | 58.97950356750648 | False | False | 0 | True | False | False | False | False | True | True | False | 1.02206966e-13 | 5.105279e-13 | 0.99998176 | 5064.985 | 5040.8657 | 5095.124 | 4.4424 | 4.4217 | 4.4671 | -0.2906 | -0.3312 | -0.2518 | 1867.0823 | 1788.1481 | 1945.3528 | 0.018 | 0.0052 | 0.037 | 0.0144 | 0.0042 | 0.0297 | 0.0076 | 0.0022 | 0.0157 | PHOENIX | 0.08328823877757062 |
| 1636148068921376768 | Gaia DR3 2051286933311580032 | 2051286933311580032 | 135441617 | 2016.0 | 290.17127999981165 | 0.04414948 | 37.73091082720331 | 0.05403776 | 0.2357502713121104 | 0.056459904 | 4.1755347 | 2.2787905 | -0.29109566200012765 | 0.055603746 | -2.260121567186946 | 0.06337706 | -0.017252762 | -0.013258554 | -0.13981561 | -0.051438995 | 0.18330099 | -0.08995912 | -0.30687386 | -0.12603593 | -0.08434859 | 0.07638833 | 414 | 0 | 412 | 2 | 0.1777575 | 428.2643 | 0.0 | 0.0 | 31 | False | 1.5031 | -- | -- | -- | -- | -- | -- | -- | 47 | 26 | 0.09099819 | 50 | 24 | 0 | 0.032246273 | 6.7542543 | 1 | 0 | 1.0054163 | 0.08898983 | 0.14224733 | 0.105735905 | 0.10415334 | -109.958275 | 77.31491 | 6.73328 | -30.988867 | False | 407 | 2336.0785755551096 | 1.5201434 | 1536.7488 | 17.266148 | 46 | 1127.3346931358217 | 8.7009325 | 129.56482 | 17.70841 | 47 | 1705.0323755571399 | 7.5450583 | 225.98001 | 16.668564 | 1.2124451 | 0 | 2 | 0 | 0 | 0 | 1.0398464 | 0.44226265 | 0.5975838 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.9037037562353 | 10.921169975727066 | 301.9260051099228 | 58.95474875157075 | False | False | 0 | True | False | False | False | False | True | True | False | 1.0224553e-13 | 5.108376e-13 | 0.99997795 | 5002.6284 | 4981.4526 | 5036.2627 | 4.602 | 4.5511 | 4.6331 | -0.8208 | -0.8784 | -0.7534 | 1583.614 | 1512.6202 | 1733.1886 | 0.0243 | 0.0074 | 0.052 | 0.0195 | 0.0059 | 0.0419 | 0.0104 | 0.0032 | 0.0225 | PHOENIX | 0.08330419708303734 |
| 1636148068921376768 | Gaia DR3 2051286967664775424 | 2051286967664775424 | 954310899 | 2016.0 | 290.1971807329532 | 0.29303434 | 37.72225240386049 | 0.3611389 | 0.5080668310750222 | 0.37153193 | 1.3674917 | 2.9852633 | -0.4817031435724317 | 0.36552507 | -2.9461430747317072 | 0.44257566 | 0.14276683 | 0.072574675 | -0.1556142 | -0.05411414 | 0.09976298 | -0.10295689 | -0.3932338 | -0.21745701 | -0.03672264 | 0.23811138 | 345 | 0 | 343 | 2 | 0.5666112 | 368.38873 | 0.0 | 0.0 | 95 | False | -- | 1.4355559 | 0.09823673 | -0.033583496 | 0.060646504 | 0.054102458 | -0.074938364 | -0.10565697 | 40 | 23 | 0.66058314 | 42 | 19 | 0 | 0.017682025 | 156.029 | 0 | 0 | 1.0209081 | 0.26176822 | 0.10280499 | 0.15333837 | 0.21728502 | -115.630585 | -75.681816 | -4.198842 | -29.578873 | False | 341 | 175.07776030520455 | 0.8039993 | 217.7586 | 20.079288 | 18 | 81.09285212765688 | 9.768502 | 8.301462 | 20.566086 | 21 | 184.31333266013976 | 7.4463835 | 24.75206 | 19.084003 | 1.5159332 | 0 | 0 | 0 | 0 | 0 | 1.4820824 | 0.48679733 | 0.99528503 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 69.9049148304103 | 10.898961260981507 | 301.95982243002953 | 58.94095155723347 | False | False | 0 | False | False | False | False | False | False | False | False | 4.541147e-10 | 2.3747997e-08 | 0.9999982 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08333124886811095 |
This query has returned very large number of columns, so we select the most useful columns to make the catalog easier to use with TweakReg.
reduced_query = gaia_query['ra', 'dec', 'phot_g_mean_mag']
reduced_query
| ra | dec | phot_g_mean_mag |
|---|---|---|
| deg | deg | mag |
| float64 | float64 | float32 |
| 290.2248611356283 | 37.80192950361359 | 18.957083 |
| 290.22327972754084 | 37.80283944795903 | 20.817099 |
| 290.22261994880597 | 37.80270752474709 | 16.63462 |
| 290.2248309701555 | 37.80058611405895 | 17.579128 |
| 290.2225602638809 | 37.80397847004457 | 18.630041 |
| 290.22681959687196 | 37.80112772770517 | 20.685486 |
| 290.2275249308142 | 37.803472775139035 | 19.896347 |
| 290.2242320409821 | 37.80039510739463 | 20.585316 |
| 290.22302939666463 | 37.8003404115494 | 18.381582 |
| ... | ... | ... |
| 290.1710542799103 | 37.874210138824154 | 18.457401 |
| 290.1228125225418 | 37.7819163631625 | 19.053267 |
| 290.1750101665401 | 37.87602842402639 | 17.952637 |
| 290.2882629014902 | 37.86913015536792 | 19.127296 |
| 290.1194354085638 | 37.80348115009846 | 19.38391 |
| 290.33007288300433 | 37.799005196546766 | 20.413414 |
| 290.14278652187744 | 37.75037525259858 | 16.866798 |
| 290.17127999981165 | 37.73091082720331 | 17.266148 |
| 290.1971807329532 | 37.72225240386049 | 20.079288 |
Then we write this catalog to an ascii file for use with TweakReg.
reduced_query.write('gaia.cat', format='ascii.commented_header', overwrite=True)
3. Align with TweakReg#
With the catalogs downloaded and the headers populated, we now need to run TweakReg with each catalog passed into the refcat parameter. The steps below compare the astrometric residuals obtained from aligning to each refcat. In each case, we set the updatehdr parameter to False to avoid altering the WCS of the images until we are satisfied with the alignment by inspecting both the TweakReg shift file and the astrometric residual plots.
3.1 SDSS Alignment#
refcat = 'sdss.cat'
wcsname = 'SDSS' # Specify the WCS name for this alignment.
cw = 3.5 # 2x the FWHM of the PSF = 3.5 pixels for ACS/WFC, WFC3/UVIS or 2.5 for WFC3/IR.
tweakreg.TweakReg('*flc.fits', # Pass the input images to tweakreg.
imagefindcfg={'threshold': 500., 'conv_width': cw}, # Detection parameters that vary for different datasets.
refcat=refcat, # Use user supplied catalog (Gaia).
interactive=False, # Don't show the interactive interface.
see2dplot=False, # Suppress additional figures in this test.
shiftfile=True, # Save output shift file to examine shifts later.
outshifts='SDSS_shifts.txt', # Name of the shift file that will be saved.
wcsname=wcsname, # Give our WCS a new name defined above.
reusename=True, # Overwrite the WCS if it exists.
ylimit=1.5,
fitgeometry='rscale', # Allow for translation, rotation, and plate scaling.
updatehdr=False) # Don't update the header with new WCS until later.
clear_output()
# If the alignment is unsuccessful then stop the notebook.
with open('SDSS_shifts.txt', 'r') as shift:
for line_number, line in enumerate(shift, start=1):
if "nan" in line:
raise ValueError('nan found in line {} in shift file'.format(line_number))
else:
continue
3.2 Inspect the shift file and fit residuals#
We can look at the shift file to see how well the fit did (or we could open the output png images for more information). The columns are:
Filename, X Shift [pixels], Y Shift [pixels], Rotation [degrees], Scale, X RMS [pixels], Y RMS [pixels]
shift_table = Table.read('SDSS_shifts.txt',
format='ascii.no_header',
names=['file', 'dx', 'dy', 'rot', 'scale', 'xrms', 'yrms'])
formats = ['.2f', '.2f', '.3f', '.5f', '.2f', '.2f']
for i, col in enumerate(shift_table.colnames[1:]):
shift_table[col].format = formats[i]
shift_table
| file | dx | dy | rot | scale | xrms | yrms |
|---|---|---|---|---|---|---|
| str18 | float64 | float64 | float64 | float64 | float64 | float64 |
| ibwb01xqq_flc.fits | 1.48 | 0.67 | 0.003 | 0.99993 | 0.37 | 0.35 |
| ibwb01xxq_flc.fits | 1.59 | -0.01 | 360.000 | 1.00011 | 0.41 | 0.53 |
| ibwb01xrq_flc.fits | 1.48 | 0.58 | 0.002 | 0.99995 | 0.37 | 0.36 |
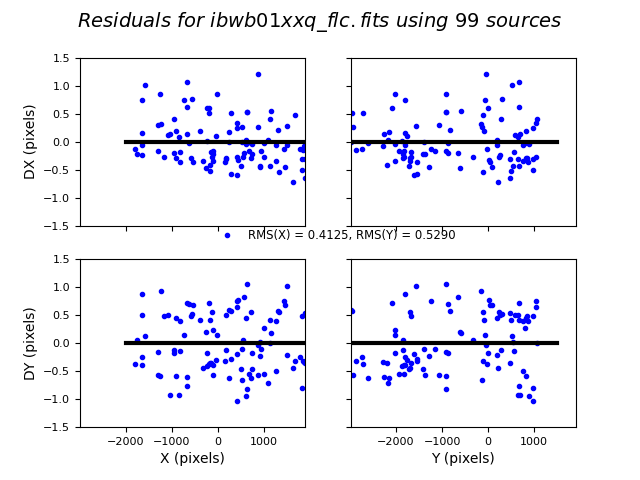
Show the astrometric residual plots.
We can see in the plots above that the RMS is fairly large at about 0.5 pixels. This is generally considered a poor fit, with the desired RMS being < 0.1 pixels depending on the number of sources. This is likely because the SDSS astrometric precision is insufficient for a high-quality HST alignment. One approach would be to align the first image to SDSS and then align the remaining HST images to one another. This would improve both the absolute and relative alignment of the individual frames.
3.3 Gaia Alignment#
refcat = 'gaia.cat'
wcsname = 'Gaia' # Specify the WCS name for this alignment.
cw = 3.5 # 2x the FWHM of the PSF = 3.5 pixels for ACS/WFC, WFC3/UVIS or 2.5 for WFC3/IR.
tweakreg.TweakReg('*flc.fits', # Pass the input images to tweakreg.
imagefindcfg={'threshold': 500., 'conv_width': cw}, # Detection parameters that vary for different datasets.
refcat=refcat, # Use user supplied catalog (Gaia).
interactive=False, # Don't show the interactive interface.
see2dplot=False, # Suppress additional figures in this test.
shiftfile=True, # Save output shift file to examine shifts later.
outshifts='Gaia_shifts.txt', # Name of the shift file that will be saved.
wcsname=wcsname, # Give our WCS a new name defined above.
reusename=True, # Overwrite the WCS if it exists.
ylimit=0.3, # The ylimit for the residuals plot.
fitgeometry='rscale', # Allow for translation, rotation, and plate scaling.
searchrad=0.1, # With many sources use a smaller search radius for convergence.
updatehdr=False) # Don't update the header with new WCS until later.
clear_output()
# If the alignment is unsuccessful then stop the notebook.
with open('Gaia_shifts.txt', 'r') as shift:
for line_number, line in enumerate(shift, start=1):
if "nan" in line:
raise ValueError('nan found in line {} in shift file'.format(line_number))
else:
continue
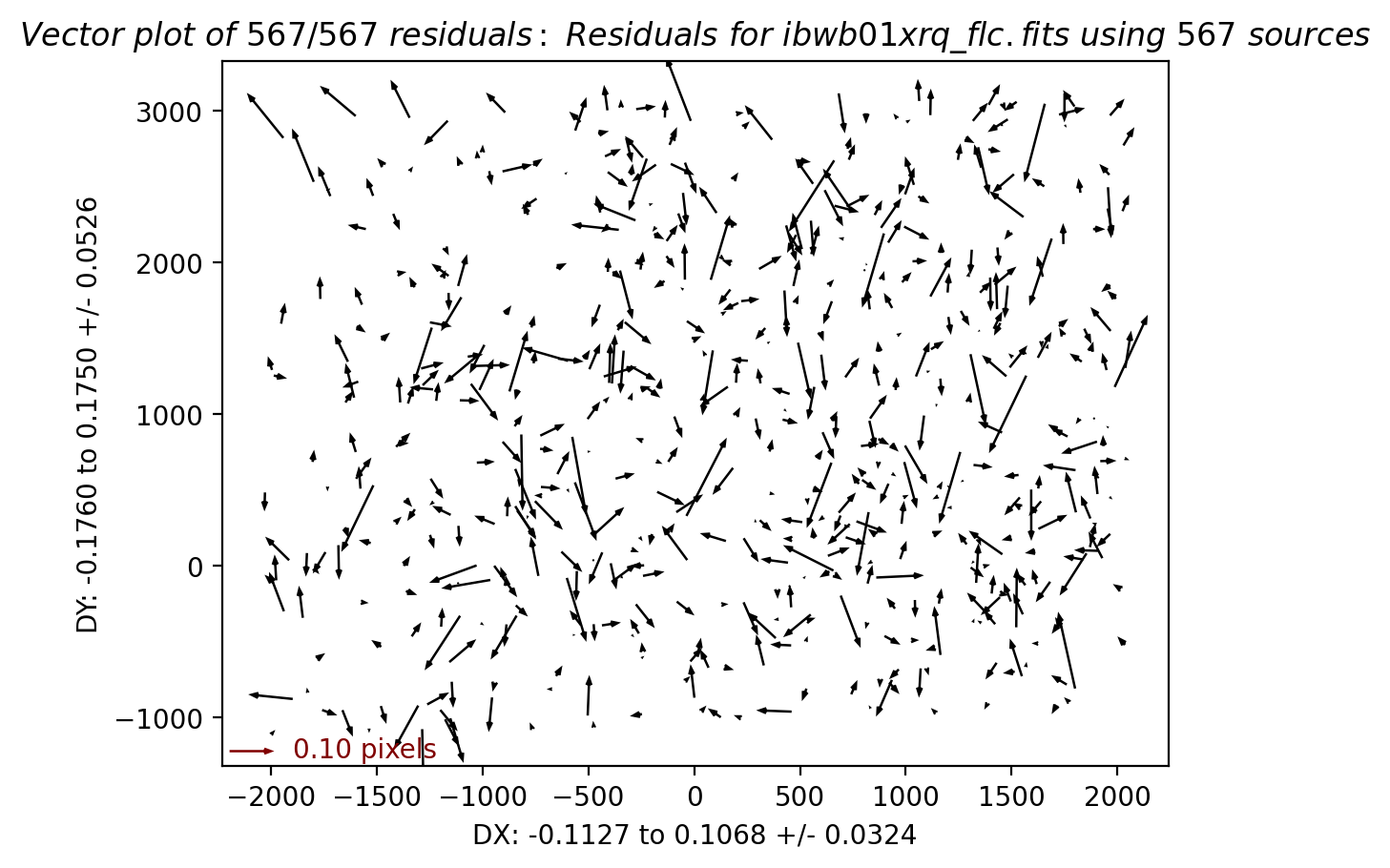
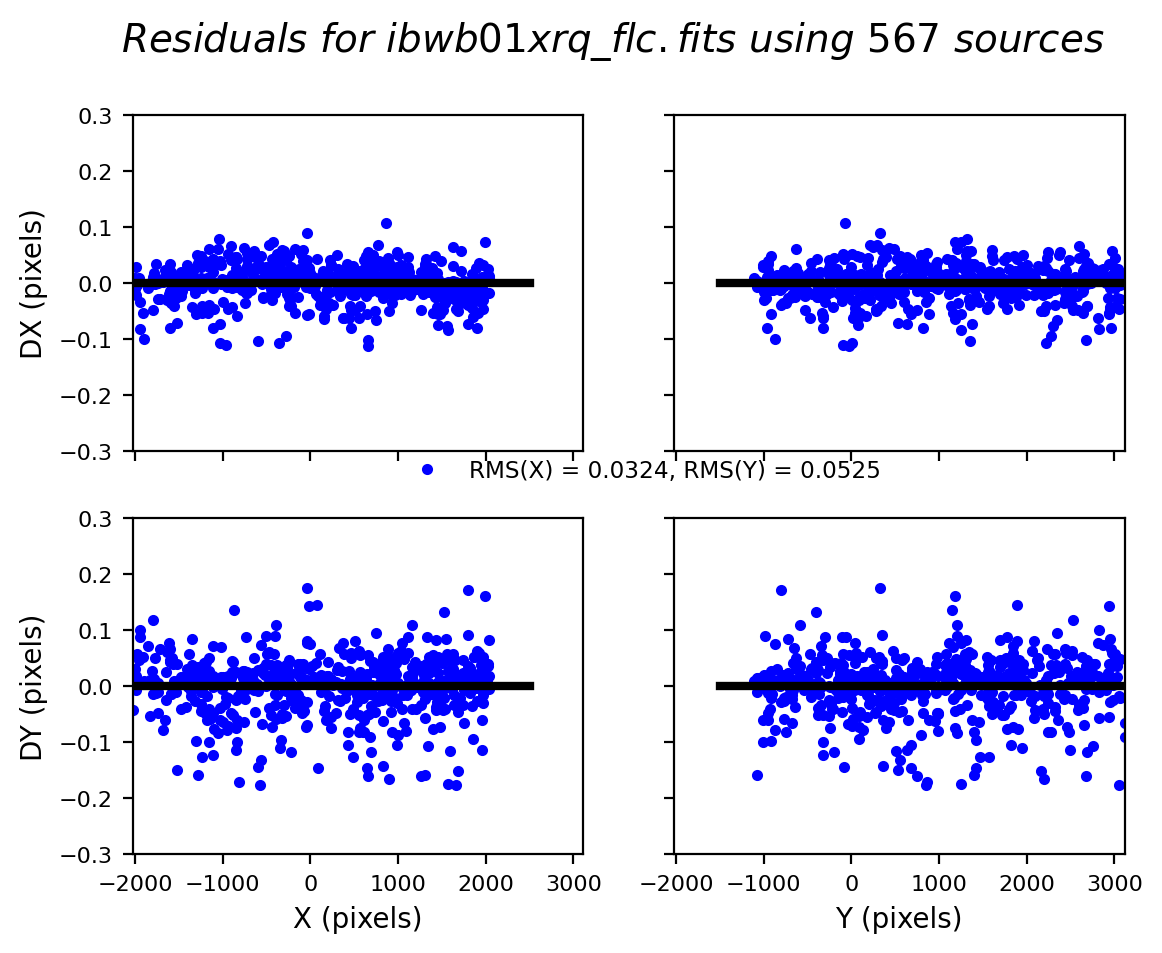
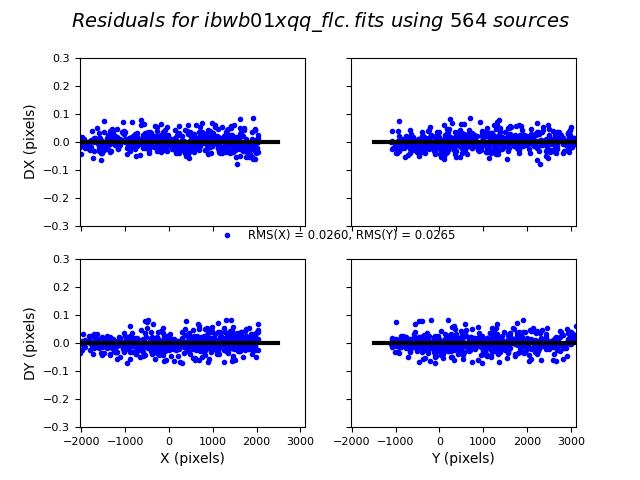
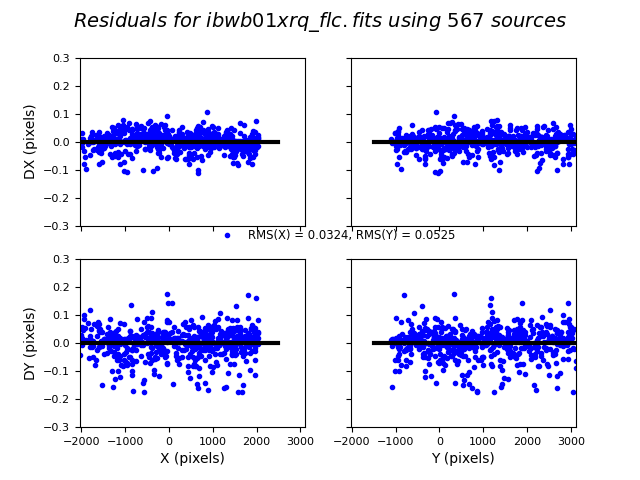
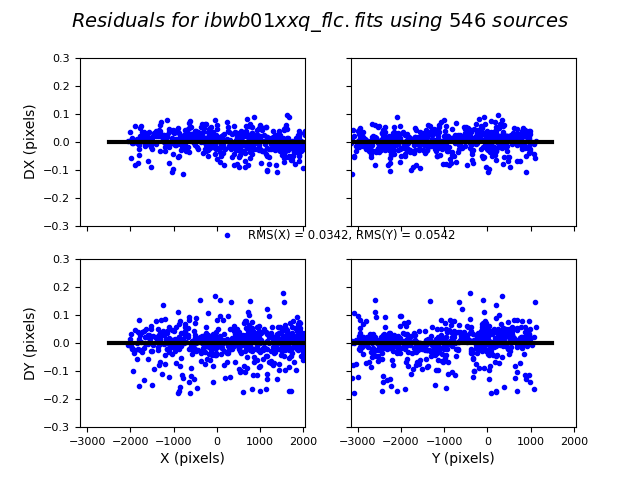
3.4 Inspect the shift file and fit residuals#
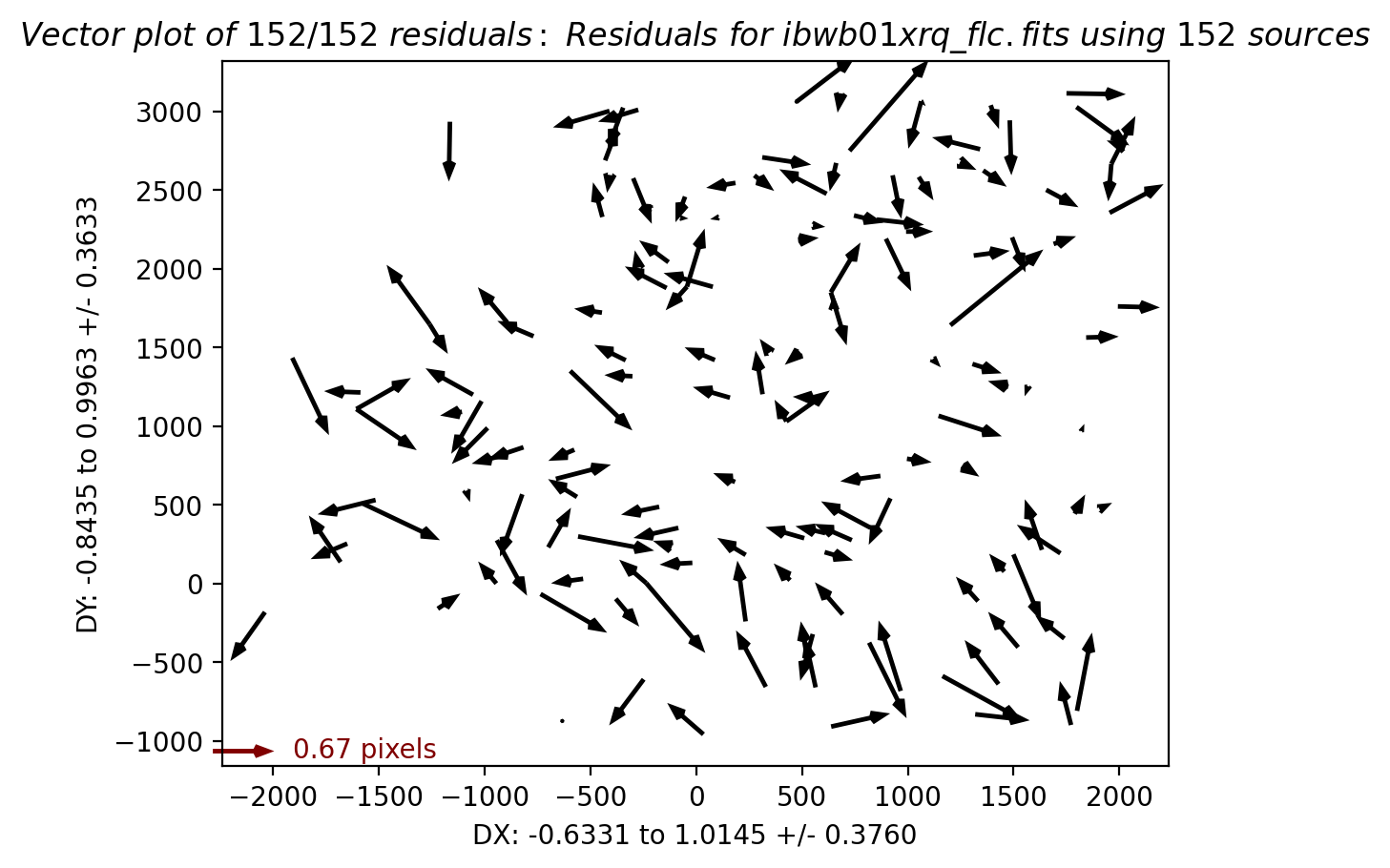
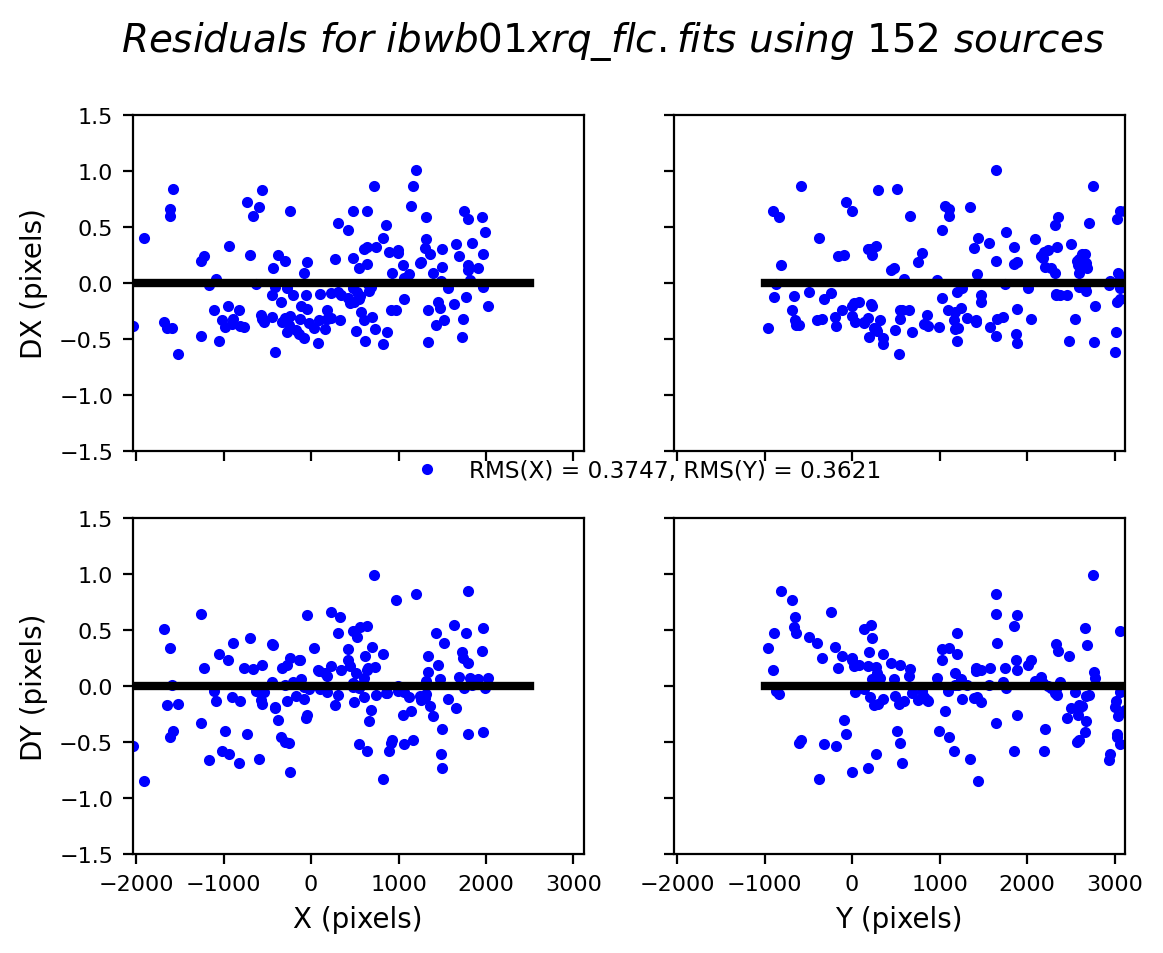
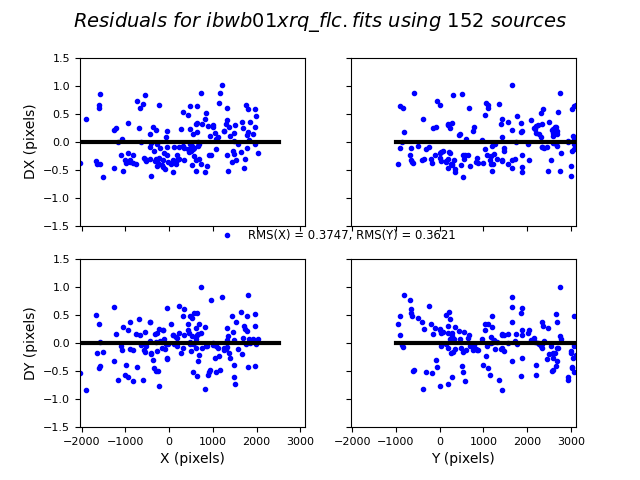
We can similarly look at the shift file from alignment to the Gaia catalog. As expected, the Gaia catalog does quite a bit better, with rms residuals ~0.05 pixels.
The columns are: Filename, X Shift [pixels], Y Shift [pixels], Rotation [degrees], Scale, X RMS [pixels], Y RMS [pixels]
shift_table = Table.read('Gaia_shifts.txt',
format='ascii.no_header',
names=['file', 'dx', 'dy', 'rot', 'scale', 'xrms', 'yrms'])
formats = ['.2f', '.2f', '.3f', '.5f', '.2f', '.2f']
for i, col in enumerate(shift_table.colnames[1:]):
shift_table[col].format = formats[i]
shift_table
| file | dx | dy | rot | scale | xrms | yrms |
|---|---|---|---|---|---|---|
| str18 | float64 | float64 | float64 | float64 | float64 | float64 |
| ibwb01xqq_flc.fits | -0.22 | 0.10 | 360.000 | 1.00000 | 0.03 | 0.03 |
| ibwb01xxq_flc.fits | -0.21 | 0.10 | 0.000 | 1.00001 | 0.03 | 0.05 |
| ibwb01xrq_flc.fits | -0.22 | 0.11 | 0.000 | 1.00000 | 0.03 | 0.05 |
Print the Number of Gaia Matches per image from the TweakReg output.
rootname1 = 'ibwb01xqq'
rootname2 = 'ibwb01xrq'
rootname3 = 'ibwb01xxq'
match_tab1 = ascii.read(rootname1+'_flc_catalog_fit.match') # load match file in astropy table
match_tab2 = ascii.read(rootname2+'_flc_catalog_fit.match') # load match file in astropy table
match_tab3 = ascii.read(rootname3+'_flc_catalog_fit.match') # load match file in astropy table
print(rootname1+': Number of Gaia Matches =', len(match_tab1))
print(rootname2+': Number of Gaia Matches =', len(match_tab2))
print(rootname3+': Number of Gaia Matches =', len(match_tab3))
ibwb01xqq: Number of Gaia Matches = 564
ibwb01xrq: Number of Gaia Matches = 567
ibwb01xxq: Number of Gaia Matches = 546
Compare the RMS and number of matches from TweakReg with the MAST alignment results.
wcstable
| filename | DETECTOR | EXPTIME | WCSNAME | NMATCHES | RMS_RA | RMS_DEC | RMS_RA_pix | RMS_DEC_pix |
|---|---|---|---|---|---|---|---|---|
| str18 | str4 | float64 | str30 | int64 | float64 | float64 | float64 | float64 |
| ibwb01xqq_flc.fits | UVIS | 30.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 435 | 2.4 | 2.3 | 0.06 | 0.06 |
| ibwb01xrq_flc.fits | UVIS | 360.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 456 | 3.6 | 3.2 | 0.09 | 0.08 |
| ibwb01xxq_flc.fits | UVIS | 360.0 | IDC_2731450pi-FIT_IMG_GAIAeDR3 | 429 | 3.9 | 3.8 | 0.1 | 0.1 |
The new Gaia solution looks better than the MAST solution, so we rerun TweakReg below and update the image headers with the new WCS solution.
For more details on absolute astrometry in HST images, see Section 4.5 in the DrizzlePac Handbook. Show the astrometric residual plots:
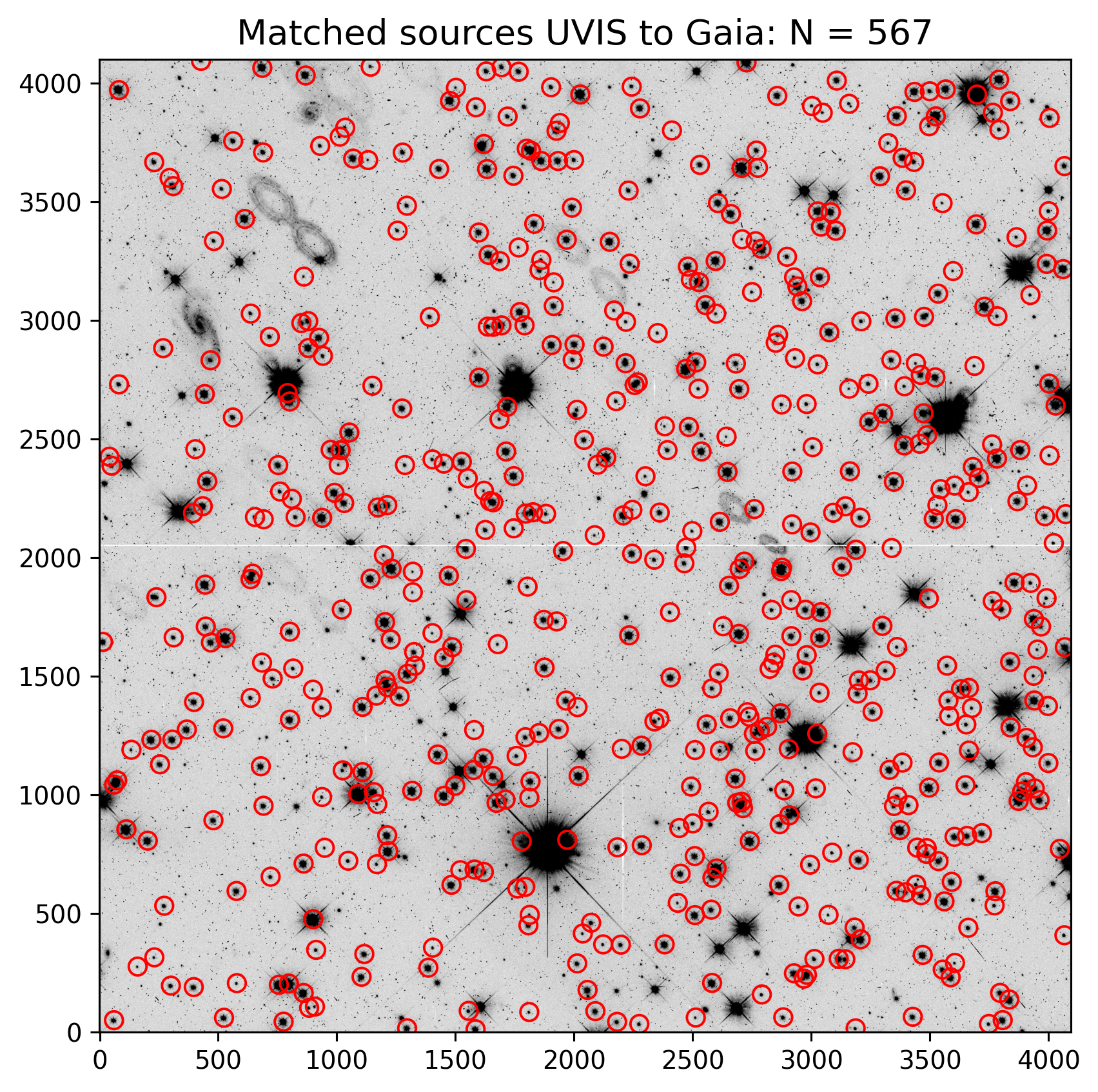
3.5 Overplot Matched Sources on the Image#
Now, let’s plot the sources that were matched between all images on the “bottom” UVIS chip. This is referred to as chip 2 or SCI, 1 or extension 1. The cell below reads in the *_fit.match file as an astropy table. Unfortunately, it doesn’t automatically name columns so we look at the header to grab the columns.
To verify that TweakReg obtained an acceptable fit between matched source catalogs, it is useful to inspect the results before updating the image header WCS. In the figure below, sources that are matched with Gaia are overplotted on one of the input FLC frames with the two chips merged together.
It can be useful to check that TweakReg locked onto stars and not hot pixels or other detector artifacts before proceeding to update the image header.
# Choose one of the files to view.
# rootname = 'ibwb01xqq'
rootname = 'ibwb01xrq'
# rootname = 'ibwb01xxq'
plt.figure(figsize=(7, 7), dpi=140)
chip1_data = fits.open(rootname+'_flc.fits')['SCI', 2].data
chip2_data = fits.open(rootname+'_flc.fits')['SCI', 1].data
fullsci = np.concatenate([chip2_data, chip1_data])
z1, z2 = ZScaleInterval().get_limits(fullsci)
plt.imshow(fullsci, cmap='Greys', origin='lower', vmin=z1, vmax=z2)
match_tab = ascii.read(rootname+'_flc_catalog_fit.match') # Load the match file in astropy table.
match_tab_chip1 = match_tab[match_tab['col15'] == 2] # Filter the table for sources on chip 1.
match_tab_chip2 = match_tab[match_tab['col15'] == 1] # Filter the table for sources on chip 2.
x_cord1, y_cord1 = match_tab_chip1['col11'], match_tab_chip1['col12']
x_cord2, y_cord2 = match_tab_chip2['col11'], match_tab_chip2['col12']
plt.scatter(x_cord1, y_cord1+2051, s=50, edgecolor='r', facecolor='None', label='Matched Sources')
plt.scatter(x_cord2, y_cord2, s=50, edgecolor='r', facecolor='None')
plt.title(f'Matched sources UVIS to Gaia: N = {len(match_tab)}', fontsize=14)
plt.show()
3.6 Update header with optimal parameters#
Once you’ve decided on the optimal set of parameters for your alignment, it is safe to update the header of the input data, (assuming that the new solution is better than the MAST alignment.) To apply these transformations to the image, we simply need to run TweakReg the same as before, but set the parameter updatehdr equal to True:
refcat = 'gaia.cat'
wcsname = 'Gaia' # Specify the WCS name for this alignment
cw = 3.5 # 2x the FWHM of the PSF = 3.5 for ACS/WFC, WFC3/UVIS or 2.5 for WFC3/IR
tweakreg.TweakReg('*flc.fits', # Pass the input images to tweakreg.
imagefindcfg={'threshold': 500., 'conv_width': cw}, # Detection parameters that vary for different datasets.
refcat=refcat, # Use user supplied catalog (Gaia).
interactive=False, # Don't show the interactive interface.
see2dplot=False, # Suppress additional figures in this test.
shiftfile=True, # Save output shift file to examine shifts later.
outshifts='Gaia_shifts.txt', # Name of the shift file that will be saved.
wcsname=wcsname, # Give our WCS a new name defined above.
reusename=True, # Overwrite the WCS if it exists.
ylimit=0.3, # The ylimit for the residuals plot.
fitgeometry='rscale', # Allow for translation, rotation, and plate scaling.
searchrad=0.1, # With many sources use a smaller search radius for convergence.
updatehdr=True) # Update the header with new WCS solution.
clear_output()
4. Combine the Images using AstroDrizzle#
While the three sets of FLC files are now aligned, in this example we drizzle together only the two long exposures. When exposures are very different lengths, drizzling them together doesn’t work well when ‘EXP’ weighting is used. For objects that saturate in the long exposures, the problem occurs at the boundary where the signal transitions from only being present in short exposures near the core to being present at larger radii in the longer exposures. The result is a discontinuity in the PSF radial profile and a resulting flux that is too low in some regions. For photometry of saturated objects, the short exposures should be drizzled separately from the long exposures.
Next, we combine the images through drizzling, and retrieve some recommended values for this process from the MDRIZTAB reference file. The parameters in this file are different for each detector and are based on the number of input frames. These are a good starting point for drizzling and may be adjusted based on your science goals.
input_images = sorted(glob.glob('ibwb01x[rx]q_flc.fits'))
mdz = fits.getval(input_images[0], 'MDRIZTAB', ext=0).split('$')[1]
print('Searching for the MDRIZTAB file:', mdz)
!crds sync --hst --files {mdz} --output-dir {os.environ['iref']}
Searching for the MDRIZTAB file: ubi1853qi_mdz.fits
CRDS - INFO - Symbolic context 'hst-latest' resolves to 'hst_1298.pmap'
CRDS - INFO - Reorganizing 0 references from 'instrument' to 'flat'
CRDS - INFO - Reorganizing from 'instrument' to 'flat' cache, removing instrument directories.
CRDS - INFO - Syncing explicitly listed files.
CRDS - INFO - 0 errors
CRDS - INFO - 0 warnings
CRDS - INFO - 4 infos
def get_vals_from_mdriztab(input_images,
kw_list=[
'driz_sep_bits',
'combine_type',
'driz_cr_snr',
'driz_cr_scale',
'final_bits']):
'Get only selected parameters from the MDRIZTAB.'
mdriz_dict = getMdriztabPars(input_images)
requested_params = {}
print('Outputting the following parameters:')
for k in kw_list:
requested_params[k] = mdriz_dict[k]
print(k, mdriz_dict[k])
return requested_params
selected_params = get_vals_from_mdriztab(input_images)
- MDRIZTAB: AstroDrizzle parameters read from row 2.
Outputting the following parameters:
driz_sep_bits 96
combine_type minmed
driz_cr_snr 3.5 3.0
driz_cr_scale 1.2 0.7
final_bits 96
Note that the parameter final_bits= ‘96’ is equivalent to final_bits= ‘64, 32’ because they are added in a bit-wise manner.
# To override any of the above values:
selected_params['driz_sep_bits'] = '256, 64, 32'
selected_params['final_bits'] = '256, 64, 32'
# selected_params['combine_type'] = 'median'
# selected_params['driz_cr_snr'] = '4.0 3.5'
# selected_params['driz_cr_scale'] = '1.5 1.2'
astrodrizzle.AstroDrizzle(input_images,
output='f606w',
preserve=False,
clean=True,
build=False,
context=False,
skymethod='match',
**selected_params)
clear_output()
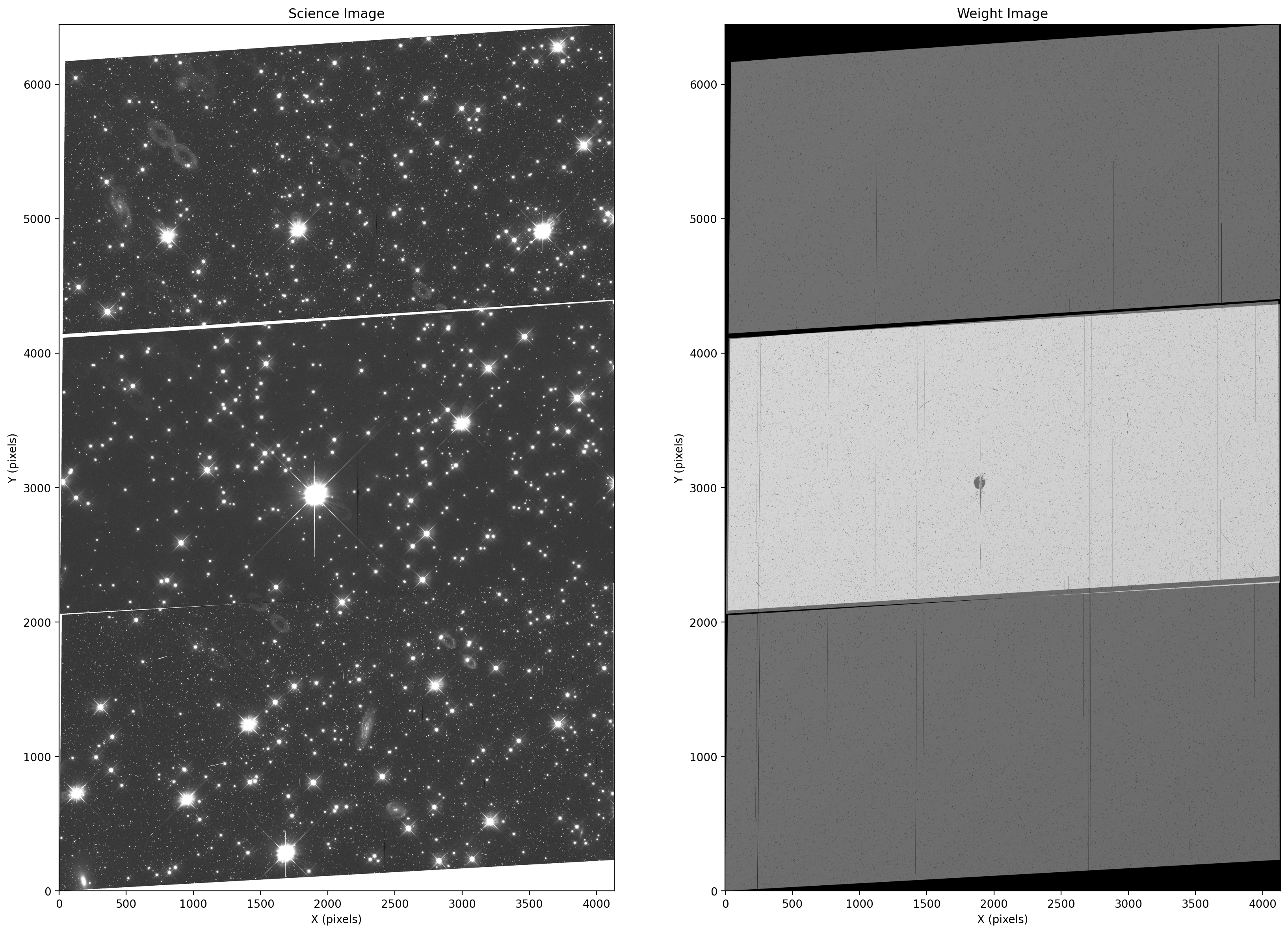
Display the combined science and weight images. In this example there appear to be three detector chips due to the large dither that was used in the observations. We note that the exposures overlapped in the center region, as evidenced by the increased value of the weight map corresponding to a longer effective exposure time. In addition, the resulting mosaic is better cleaned of artifacts and cosmic rays in this region due to the multiple exposures, as seen by the uniform background in the science mosaic.
sci = fits.getdata('f606w_drc_sci.fits')
wht = fits.getdata('f606w_drc_wht.fits')
fig = plt.figure(figsize=(20, 20))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(sci, vmin=-0.05, vmax=0.4, cmap='Greys_r', origin='lower')
ax2.imshow(wht, vmin=0.0, vmax=1000, cmap='Greys_r', origin='lower')
ax1.title.set_text('Science Image')
ax2.title.set_text('Weight Image')
ax1.set_xlabel('X (pixels)')
ax1.set_ylabel('Y (pixels)')
ax2.set_xlabel('X (pixels)')
ax2.set_ylabel('Y (pixels)')
For more details on inspecting the drizzled products after reprocessing, see Section 7.3 in the DrizzlePac Handbook.
Conclusions#
Many other services have interfaces for querying catalogs that can also be used to align HST images. In general, Gaia works very well for HST due to its high precision, but can have a low number of sources in some regions, especially at high galactic latitudes. Aligning images to an absolute frame provides a way to make data comparable across many epochs/detectors/observatories, and in many cases, makes the alignment easier to complete.
About this Notebook#
Created: 14 Dec 2018; V. Bajaj
Updated: 27 May 2025; M. Revalski, V. Bajaj, & J. Mack
Source: GitHub spacetelescope/hst_notebooks
Additional Resources#
Below are some additional resources that may be helpful. Please send any questions through the HST Help Desk, selecting the DrizzlePac category.
Citations#
If you use Python packages such as astropy, astroquery, drizzlepac, matplotlib, or numpy for published research, please cite the authors.
Follow these links for more information about citing various packages: