
Masking Persistence in WFC3/IR Images#
Learning Goals#
This notebook shows how to use the Hubble Space Telescope WFC3/IR persistence model to flag pixels affected by persistence in the calibrated (FLT) science images. When the images are sufficiently dithered to step over the observed persistence artifacts, AstroDrizzle may be used to exclude those flagged pixels when combining the FLT frames.
By the end of this tutorial, you will:
Download images and persistence products from MAST.
Flag affected pixels in the data quality arrays of the FLT images.
Redrizzle the FLT images to produce a “clean” DRZ combined product.
Table of Contents#
Introduction
1. Imports
2. Data
4. Conclusions
Additional Resources
About this Notebook
Citations
Introduction #
Image persistence in the IR array occurs whenever a pixel is exposed to light that exceeds more than about half of the full well of a pixel in the array. Persistence can occur within a single visit, as the different exposures in a visit are dithered. Persistence also occurs from observations in a previous visit of completely different fields.
Image persistence is seen in a small, but non-negligible fraction of WFC3/IR exposures. Its properties are discussed in in Section 5.7.9 of the WFC3 Instrument Handbook and in Chapter 8 of the WFC3 Data Handbook. Persistence is primarily a function of the degree to which a pixel is filled (in electrons) and the time since this occurred. Additional information is available from the WFC3 Persistence Webpage.
As described in Section 8.3 of the Data Handbook, there are two possible ways to mitigate persistence: 1.) exclude the affected pixels from the analysis or 2.) subtract the persistence model directly from the image.
This notebook illustrates the first method and shows how to use the model to flag affected pixels in the data quality (DQ) array of each FLT image. When the images are sufficiently dithered, affected regions of the detector may be replaced with ‘good’ pixels from other exposures in the visit when combining the exposures with AstroDrizzle. Note that this reduces the effective exposure time in those regions of the combined image.
In the second method, the persistence-corrected FLT frames, downloaded in Section 2.2 of this notebook, may be used directly for analysis. Alternatively, a scaled version of the persistence model may be subtracted from each FLT image until an adequate correction is achieved. In this case, flagging the affected pixels in the DQ arrays would not be required.
1. Imports #
This notebook assumes you have installed the required libraries as described here.
We import:
glob for finding lists of files
os for setting environment variables
shutil for managing directories
urllib for obtaining the Persistence products from MAST
matplotlib.pyplot for plotting data
astropy.io fits for accessing FITS files
astroquery for downloading data from MAST
ccdproc for querying keyword values in the FITS headers
drizzlepac astrodrizzle for combining images
import glob
import os
import shutil
import urllib
import matplotlib.pyplot as plt
from astropy.io import fits
from astroquery.mast import Observations
from ccdproc import ImageFileCollection
from drizzlepac import astrodrizzle
2. Data #
2.1 Download the WFC3/IR observations from MAST #
Here, we obtain WFC3/IR observations from the Grism Lens-Amplified Survey from Space (GLASS) program 13459, Visit 29 in the F140W filter. These exposures were impacted by persistence from grism G102 exposures obtained just prior to these.
The following commands query the Mikulski Archive for Space Telescopes (MAST) and then download the FLT and DRZ data products to the current directory.
data_list = Observations.query_criteria(obs_id="ica529*", filters="F140W")
Observations.download_products(
data_list["obsid"],
mrp_only=False,
download_dir="./data",
productSubGroupDescription=["FLT", "DRZ"],
)
science_files = glob.glob("data/mastDownload/HST/*/*fits")
for im in science_files:
filename = os.path.basename(im)
new_path = os.path.join(".", filename)
os.rename(im, new_path)
data_directory = "./data"
try:
if os.path.isdir(data_directory):
shutil.rmtree(data_directory)
except Exception as e:
print(f"An error occured while deleting the directory {data_directory}: {e}")
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ica529rmq_flt.fits to ./data/mastDownload/HST/ica529rmq/ica529rmq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ica529rsq_flt.fits to ./data/mastDownload/HST/ica529rsq/ica529rsq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ica529s0q_flt.fits to ./data/mastDownload/HST/ica529s0q/ica529s0q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ica529s6q_flt.fits to ./data/mastDownload/HST/ica529s6q/ica529s6q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ica529030_drz.fits to ./data/mastDownload/HST/ica529030/ica529030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529s6_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529s6/hst_13459_29_wfc3_ir_f140w_ica529s6_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529s6_flt.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529s6/hst_13459_29_wfc3_ir_f140w_ica529s6_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529rs_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529rs/hst_13459_29_wfc3_ir_f140w_ica529rs_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529rs_flt.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529rs/hst_13459_29_wfc3_ir_f140w_ica529rs_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529rm_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529rm/hst_13459_29_wfc3_ir_f140w_ica529rm_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529rm_flt.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529rm/hst_13459_29_wfc3_ir_f140w_ica529rm_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529s0_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529s0/hst_13459_29_wfc3_ir_f140w_ica529s0_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529s0_flt.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529s0/hst_13459_29_wfc3_ir_f140w_ica529s0_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_f140w_ica529_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_f140w_ica529/hst_13459_29_wfc3_ir_f140w_ica529_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/hst_13459_29_wfc3_ir_total_ica529_drz.fits to ./data/mastDownload/HST/hst_13459_29_wfc3_ir_total_ica529/hst_13459_29_wfc3_ir_total_ica529_drz.fits ...
[Done]
This Visit contains 4 consecutive dithered FLT exposures in the F140W filter, which are obtained in a single orbit. The following commands print the values of keywords describing those data, where the POSTARG* values represent the commanded x-axis and y-axis offsets in arcseconds.
image_collection = ImageFileCollection(
"./",
keywords=[
"asn_id",
"targname",
"filter",
"samp_seq",
"nsamp",
"exptime",
"postarg1",
"postarg2",
"date-obs",
"time-obs",
],
glob_include="ica529*flt.fits",
ext=0,
)
try:
summary_table = image_collection.summary
if summary_table:
print(summary_table)
else:
print("No FITS files matched the pattern or no relevant data found.")
except Exception as e:
print(f"An error occurred while creating the summary table: {e}")
file asn_id targname ... postarg2 date-obs time-obs
------------------ --------- --------------- ... -------- ---------- --------
ica529rmq_flt.fits ICA529030 MACS0717.5+3745 ... 0.0 2014-02-19 11:26:12
ica529rsq_flt.fits ICA529030 MACS0717.5+3745 ... 0.424 2014-02-19 11:47:06
ica529s0q_flt.fits ICA529030 MACS0717.5+3745 ... 1.212 2014-02-19 12:55:38
ica529s6q_flt.fits ICA529030 MACS0717.5+3745 ... 0.788 2014-02-19 13:16:32
2.2 Download the persistence model products #
To find the URL of the tar file containing the persistence fits files, visit the PERSIST Search Form and search for dataset=’ica529*’.
You may hover over the link in the Visit column to get the URL to the gzipped tar file for Visit 29. This URL is called in the Python code below. The persistence model data products we will use to create a mask are named ‘rootname_persist.fits’ and contain any contributions from either external or internal persistence.
External persistence is defined as residual signal that is generated by an earlier visit, and internal persistence as that generated within the same visit as the image in question. External persistence typically comes from a prior scheduled WFC3/IR program and is not within the control of the observer. Internal persistence can be mitigated by the observer by dithering the exposures within a given visit.
This cell may take several minutes to complete, as it will download persistence products for all images in the visit, and not just those for the F140W filter.
url = "https://archive.stsci.edu/pub/wfc3_persist/13459/Visit29/13459.Visit29.tar.gz"
filename = "13459.Visit29.tar.gz"
try:
with urllib.request.urlopen(url) as response:
with open(filename, "wb") as out_file:
out_file.write(response.read())
print("Extracting files....")
# Extract files:
!tar -zxvf {filename}
# Remove the files after extraction
os.remove("13459.Visit29.tar.gz")
except Exception as e:
print(f"An error occured: {e}")
Extracting files....
13459.Visit29/
13459.Visit29/ica529rmq_extper.fits
13459.Visit29/ica529rmq_flt_cor.fits
13459.Visit29/ica529rmq_persist.fits
13459.Visit29/ica529roq_extper.fits
13459.Visit29/ica529roq_flt_cor.fits
13459.Visit29/ica529roq_persist.fits
13459.Visit29/ica529rsq_extper.fits
13459.Visit29/ica529rsq_flt_cor.fits
13459.Visit29/ica529rsq_persist.fits
13459.Visit29/ica529rvq_extper.fits
13459.Visit29/ica529rvq_flt_cor.fits
13459.Visit29/ica529rvq_persist.fits
13459.Visit29/ica529s0q_extper.fits
13459.Visit29/ica529s0q_flt_cor.fits
13459.Visit29/ica529s0q_persist.fits
13459.Visit29/ica529s2q_extper.fits
13459.Visit29/ica529s2q_flt_cor.fits
13459.Visit29/ica529s2q_persist.fits
13459.Visit29/ica529s6q_extper.fits
13459.Visit29/ica529s6q_flt_cor.fits
13459.Visit29/ica529s6q_persist.fits
13459.Visit29/ica529s9q_extper.fits
13459.Visit29/ica529s9q_flt_cor.fits
13459.Visit29/ica529s9q_persist.fits
13459.Visit29/ica529seq_extper.fits
13459.Visit29/ica529seq_flt_cor.fits
13459.Visit29/ica529seq_persist.fits
13459.Visit29/ica529sgq_extper.fits
13459.Visit29/ica529sgq_flt_cor.fits
13459.Visit29/ica529sgq_persist.fits
13459.Visit29/ica529skq_extper.fits
13459.Visit29/ica529skq_flt_cor.fits
13459.Visit29/ica529skq_persist.fits
13459.Visit29/ica529snq_extper.fits
13459.Visit29/ica529snq_flt_cor.fits
13459.Visit29/ica529snq_persist.fits
13459.Visit29/ica529ssq_extper.fits
13459.Visit29/ica529ssq_flt_cor.fits
13459.Visit29/ica529ssq_persist.fits
13459.Visit29/ica529suq_extper.fits
13459.Visit29/ica529suq_flt_cor.fits
13459.Visit29/ica529suq_persist.fits
13459.Visit29/ica529syq_extper.fits
13459.Visit29/ica529syq_flt_cor.fits
13459.Visit29/ica529syq_persist.fits
13459.Visit29/ica529t1q_extper.fits
13459.Visit29/ica529t1q_flt_cor.fits
13459.Visit29/ica529t1q_persist.fits
This creates a subdirectory named 13459.Visit29/ in the working directory with the following products for each calibrated ‘flt.fits’ image:
persist.fits: The persistence model, including both internal and external persistence
extper.fits: The persistence model, including only external persistence
flt_corr.fits: The corrected FLT image, equal to the difference between the image and the model (‘flt.fits’ - ‘persist.fits’).
Note that limitations in the accuracy of the model can result in corrected FLT images in which persistence is not properly removed. This is especially true when prior observations were obtained in scanning mode, where the model significantly underestimates the level of persistence. In this case the model may be iteratively scaled and subtracted from the FLT frame until the residual signal is fully removed.
Alternately, this notebook shows how the user can flag the impacted pixels in the FLT data quality array then reprocess with AstroDrizzle to improve the combined image.
3. Analysis #
3.1 Display the images #
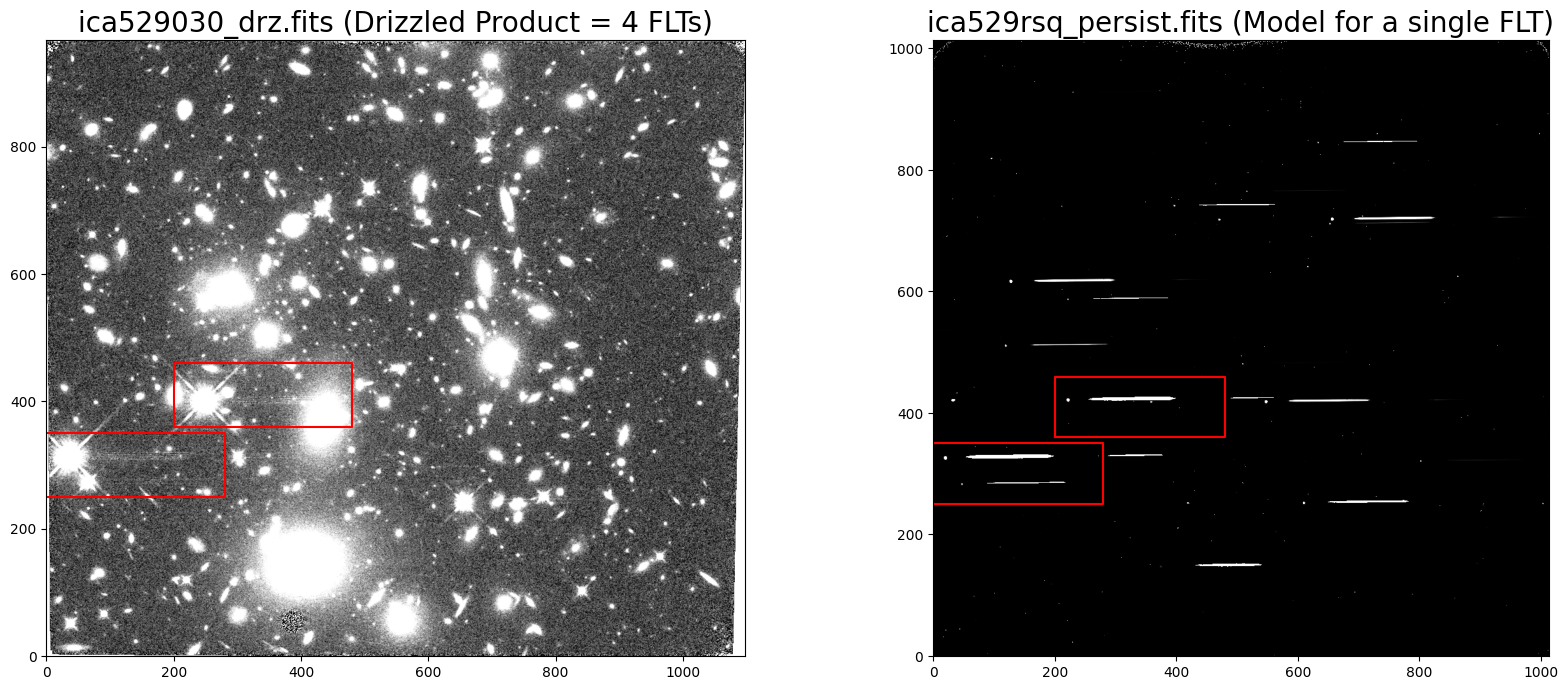
The drizzeled (DRZ) product combines the 4 individual FLT exposures and shows faint residual persistence from grism observations obtained just prior to these data. An example of the ‘Persistence Removal Evaluation’ for the first dataset ica529rmq may be found here.
The DRZ pipeline product is shown on the left, and we can see that the persistence was partially, but not completely, removed by the cosmic-ray rejection functionality in AstroDrizzle. The persistence model for the first FLT dataset is shown on the right. A pair of red boxes are overplotted on each image to highlight regions of the detector with the most visible grism persistence in the combined DRZ image.
drz = fits.getdata('ica529030_drz.fits', ext=1)
per1 = fits.getdata('13459.Visit29/ica529rsq_persist.fits', ext=1)
fig = plt.figure(figsize=(20, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(drz, vmin=0.85, vmax=1.4, cmap='Greys_r', origin='lower')
ax2.imshow(per1, vmin=0.0, vmax=0.005, cmap='Greys_r', origin='lower')
ax1.set_title('ica529030_drz.fits (Drizzled Product = 4 FLTs)', fontsize=20)
ax2.set_title('ica529rsq_persist.fits (Model for a single FLT)', fontsize=20)
ax1.plot([0, 280, 280, 0, 0], [250, 250, 350, 350, 250], c='red')
ax2.plot([0, 280, 280, 0, 0], [250, 250, 350, 350, 250], c='red')
ax1.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
ax2.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
[<matplotlib.lines.Line2D at 0x7f08d739a490>]
3.2 Use the persistence model to add DQ flags #
For any pixels in the model with a signal greater than 0.005 e-/sec, we add a flag of 16384 to the current DQ array values in each FLT frame. This threshold is flexible and should be determined empirically by the user based on the science objective and the fraction of pixels impacted. Note that the IR dark rate is 0.049 e-/s, so a threshold of 0.05 e-/s or 0.01 e-/s may be a more reasonable starting value in order to avoid flagging more pixels than can be filled in with the associated dithered FLT frames.
flt1_dq = fits.open('ica529rmq_flt.fits', mode='update')
per1 = fits.getdata('13459.Visit29/ica529rmq_persist.fits', ext=1)
flt2_dq = fits.open('ica529rsq_flt.fits', mode='update')
per2 = fits.getdata('13459.Visit29/ica529rsq_persist.fits', ext=1)
flt3_dq = fits.open('ica529s0q_flt.fits', mode='update')
per3 = fits.getdata('13459.Visit29/ica529s0q_persist.fits', ext=1)
flt4_dq = fits.open('ica529s6q_flt.fits', mode='update')
per4 = fits.getdata('13459.Visit29/ica529s6q_persist.fits', ext=1)
flt1_dq[3].data[per1 > 0.005] += 16384
flt2_dq[3].data[per2 > 0.005] += 16384
flt3_dq[3].data[per3 > 0.005] += 16384
flt4_dq[3].data[per4 > 0.005] += 16384
flt1_dq.close()
flt2_dq.close()
flt3_dq.close()
flt4_dq.close()
3.3 Redrizzle the FLT data and apply the new DQ flags #
Now, we recombine the FLT data with AstroDrizzle using the updated DQ arrays and compare with the pipeline DRZ data products. The following cell uses the default parameter values recommended for the IR detector, where final_bits tells AstroDrizzle which DQ flags to ignore (e.g. to treat as good data). All other flagged (non-zero) values will be treated as bad pixels and excluded from the combined image.
astrodrizzle.AstroDrizzle('ica529*flt.fits', output='ica529030_pcorr',
preserve=False, build=True, clean=True,
skymethod='match', sky_bits='16',
driz_sep_bits='512,16', combine_type='median',
driz_cr_snr='5.0 4.0', driz_cr_scale='3.0 2.4',
final_bits='512,16', num_cores=1)
INFO:drizzlepac.util:Setting up logfile : astrodrizzle.log
Setting up logfile : astrodrizzle.log
INFO:drizzlepac.astrodrizzle:AstroDrizzle log file: astrodrizzle.log
AstroDrizzle log file: astrodrizzle.log
INFO:drizzlepac.astrodrizzle:AstroDrizzle Version 3.10.0 started at: 20:16:42.994 (02/12/2025)
AstroDrizzle Version 3.10.0 started at: 20:16:42.994 (02/12/2025)
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:Version Information
INFO:drizzlepac.astrodrizzle:--------------------
INFO:drizzlepac.astrodrizzle:Python Version 3.11.14 | packaged by conda-forge | (main, Oct 22 2025, 22:46:25) [GCC 14.3.0]
INFO:drizzlepac.astrodrizzle:numpy Version -> 2.3.5
INFO:drizzlepac.astrodrizzle:astropy Version -> 7.2.0
INFO:drizzlepac.astrodrizzle:stwcs Version -> 1.7.5
INFO:drizzlepac.astrodrizzle:photutils Version -> 2.3.0
INFO:drizzlepac.util:==== Processing Step Initialization started at 20:16:43.000 (02/12/2025)
==== Processing Step Initialization started at 20:16:43.000 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.processInput:Executing serially
INFO:drizzlepac.processInput:Setting up output name: ica529030_pcorr_drz.fits
INFO:drizzlepac.processInput:-Creating imageObject List as input for processing steps.
INFO:drizzlepac.imageObject:Reading in MDRIZSKY of 0.1885567023501458
INFO:drizzlepac.imageObject:Reading in MDRIZSKY of 0.0
INFO:drizzlepac.imageObject:Reading in MDRIZSKY of 0.3307531652665684
INFO:drizzlepac.imageObject:Reading in MDRIZSKY of 0.02448945831326
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ica529rmq_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ica529rsq_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ica529s0q_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ica529s6q_flt.fits[DQ,1]
INFO:drizzlepac.processInput:-Creating output WCS.
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 109.39116177250045 37.74673386265051
CRVAL : 109.39116177250045 37.74673386265051
INFO:astropy.wcs.wcs:CRPIX : 549.0 484.0
CRPIX : 549.0 484.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
INFO:astropy.wcs.wcs:NAXIS : 1098 968
NAXIS : 1098 968
INFO:drizzlepac.processInput:********************************************************************************
********************************************************************************
INFO:drizzlepac.processInput:*
*
INFO:drizzlepac.processInput:* Estimated memory usage: up to 20 Mb.
* Estimated memory usage: up to 20 Mb.
INFO:drizzlepac.processInput:* Output image size: 1098 X 968 pixels.
* Output image size: 1098 X 968 pixels.
INFO:drizzlepac.processInput:* Output image file: ~ 12 Mb.
* Output image file: ~ 12 Mb.
INFO:drizzlepac.processInput:* Cores available: 1
* Cores available: 1
INFO:drizzlepac.processInput:*
*
INFO:drizzlepac.processInput:********************************************************************************
********************************************************************************
INFO:drizzlepac.util:==== Processing Step Initialization finished at 20:16:43.541 (02/12/2025)
==== Processing Step Initialization finished at 20:16:43.541 (02/12/2025)
INFO:drizzlepac.astrodrizzle:USER INPUT PARAMETERS common to all Processing Steps:
INFO:drizzlepac.astrodrizzle: build : True
INFO:drizzlepac.astrodrizzle: coeffs : True
INFO:drizzlepac.astrodrizzle: context : True
INFO:drizzlepac.astrodrizzle: crbit : 4096
INFO:drizzlepac.astrodrizzle: group :
INFO:drizzlepac.astrodrizzle: in_memory : False
INFO:drizzlepac.astrodrizzle: input : ica529*flt.fits
INFO:drizzlepac.astrodrizzle: mdriztab : False
INFO:drizzlepac.astrodrizzle: num_cores : 1
INFO:drizzlepac.astrodrizzle: output : ica529030_pcorr
INFO:drizzlepac.astrodrizzle: proc_unit : native
INFO:drizzlepac.astrodrizzle: resetbits : 4096
INFO:drizzlepac.astrodrizzle: rules_file :
INFO:drizzlepac.astrodrizzle: runfile : astrodrizzle.log
INFO:drizzlepac.astrodrizzle: stepsize : 10
INFO:drizzlepac.astrodrizzle: updatewcs : False
INFO:drizzlepac.astrodrizzle: wcskey :
INFO:drizzlepac.util:==== Processing Step Static Mask started at 20:16:43.55 (02/12/2025)
==== Processing Step Static Mask started at 20:16:43.55 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.staticMask:USER INPUT PARAMETERS for Static Mask Step:
INFO:drizzlepac.staticMask: static : True
INFO:drizzlepac.staticMask: static_sig : 4.0
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.201202; rms = 0.224134; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.006969; rms = 0.192982; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.311748; rms = 0.229180; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.027247; rms = 0.193885; static_sig = 4.00
INFO:drizzlepac.staticMask:Saving static mask to disk: ./ica529030_1014x1014_1_staticMask.fits
INFO:drizzlepac.util:==== Processing Step Static Mask finished at 20:16:43.623 (02/12/2025)
==== Processing Step Static Mask finished at 20:16:43.623 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Subtract Sky started at 20:16:43.624 (02/12/2025)
==== Processing Step Subtract Sky started at 20:16:43.624 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.sky:USER INPUT PARAMETERS for Sky Subtraction Step:
INFO:drizzlepac.sky: sky_bits : 16
INFO:drizzlepac.sky: skyclip : 5
INFO:drizzlepac.sky: skyfile :
INFO:drizzlepac.sky: skylower : None
INFO:drizzlepac.sky: skylsigma : 4.0
INFO:drizzlepac.sky: skymask_cat :
INFO:drizzlepac.sky: skymethod : match
INFO:drizzlepac.sky: skystat : median
INFO:drizzlepac.sky: skysub : True
INFO:drizzlepac.sky: skyupper : None
INFO:drizzlepac.sky: skyuser :
INFO:drizzlepac.sky: skyusigma : 4.0
INFO:drizzlepac.sky: skywidth : 0.1
INFO:drizzlepac.sky: use_static : True
INFO:stsci.skypac.utils:***** skymatch started on 2025-12-02 20:16:43.686201
***** skymatch started on 2025-12-02 20:16:43.686201
INFO:stsci.skypac.utils: Version 1.0.11
Version 1.0.11
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:'skymatch' task will apply computed sky differences to input image file(s).
'skymatch' task will apply computed sky differences to input image file(s).
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
INFO:stsci.skypac.utils:'MDRIZSKY' header keyword will represent sky value *computed* from data.
'MDRIZSKY' header keyword will represent sky value *computed* from data.
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- User specified keywords: -----
----- User specified keywords: -----
INFO:stsci.skypac.utils: Sky Value Keyword: 'MDRIZSKY'
Sky Value Keyword: 'MDRIZSKY'
INFO:stsci.skypac.utils: Data Units Keyword: 'BUNIT'
Data Units Keyword: 'BUNIT'
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Input file list: -----
----- Input file list: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ica529rmq_flt.fits'
** Input image: 'ica529rmq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ica529rmq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ica529rmq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ica529rsq_flt.fits'
** Input image: 'ica529rsq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ica529rsq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ica529rsq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ica529s0q_flt.fits'
** Input image: 'ica529s0q_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ica529s0q_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ica529s0q_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ica529s6q_flt.fits'
** Input image: 'ica529s6q_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ica529s6q_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ica529s6q_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Sky statistics parameters: -----
----- Sky statistics parameters: -----
INFO:stsci.skypac.utils: statistics function: 'median'
statistics function: 'median'
INFO:stsci.skypac.utils: lower = None
lower = None
INFO:stsci.skypac.utils: upper = None
upper = None
INFO:stsci.skypac.utils: nclip = 5
nclip = 5
INFO:stsci.skypac.utils: lsigma = 4.0
lsigma = 4.0
INFO:stsci.skypac.utils: usigma = 4.0
usigma = 4.0
INFO:stsci.skypac.utils: binwidth = 0.1
binwidth = 0.1
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Data->Brightness conversion parameters for input files: -----
----- Data->Brightness conversion parameters for input files: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ica529rmq_flt.fits
* Image: ica529rmq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.797431635711504
Conversion factor (data->brightness): 60.797431635711504
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ica529rsq_flt.fits
* Image: ica529rsq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.797431635711504
Conversion factor (data->brightness): 60.797431635711504
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ica529s0q_flt.fits
* Image: ica529s0q_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.797431635711504
Conversion factor (data->brightness): 60.797431635711504
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ica529s6q_flt.fits
* Image: ica529s6q_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.797431635711504
Conversion factor (data->brightness): 60.797431635711504
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Computing differences in sky values in overlapping regions: -----
----- Computing differences in sky values in overlapping regions: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ica529rmq_flt.fits['SCI',1]' SKY = 11.7179 [brightness units]
* Image 'ica529rmq_flt.fits['SCI',1]' SKY = 11.7179 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0.192738
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.192738
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ica529rsq_flt.fits['SCI',1]' SKY = 0 [brightness units]
* Image 'ica529rsq_flt.fits['SCI',1]' SKY = 0 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0
- EXT = 'SCI',1 delta(MDRIZSKY) = 0
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ica529s0q_flt.fits['SCI',1]' SKY = 19.8763 [brightness units]
* Image 'ica529s0q_flt.fits['SCI',1]' SKY = 19.8763 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0.326927
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.326927
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ica529s6q_flt.fits['SCI',1]' SKY = 1.8091 [brightness units]
* Image 'ica529s6q_flt.fits['SCI',1]' SKY = 1.8091 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0.0297562
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.0297562
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:***** skymatch ended on 2025-12-02 20:16:44.661486
***** skymatch ended on 2025-12-02 20:16:44.661486
INFO:stsci.skypac.utils:TOTAL RUN TIME: 0:00:00.975285
TOTAL RUN TIME: 0:00:00.975285
INFO:drizzlepac.util:==== Processing Step Subtract Sky finished at 20:16:44.712 (02/12/2025)
==== Processing Step Subtract Sky finished at 20:16:44.712 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Separate Drizzle started at 20:16:44.71 (02/12/2025)
==== Processing Step Separate Drizzle started at 20:16:44.71 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.adrizzle:Interpreted paramDict with single=True as:
{'build': True, 'stepsize': 10, 'coeffs': True, 'wcskey': '', 'kernel': 'turbo', 'wt_scl': 'exptime', 'pixfrac': 1.0, 'fillval': None, 'bits': 528, 'compress': False, 'units': 'cps'}
INFO:drizzlepac.adrizzle:USER INPUT PARAMETERS for Separate Drizzle Step:
INFO:drizzlepac.adrizzle: bits : 528
INFO:drizzlepac.adrizzle: build : False
INFO:drizzlepac.adrizzle: clean : True
INFO:drizzlepac.adrizzle: coeffs : True
INFO:drizzlepac.adrizzle: compress : False
INFO:drizzlepac.adrizzle: crbit : None
INFO:drizzlepac.adrizzle: fillval : None
INFO:drizzlepac.adrizzle: kernel : turbo
INFO:drizzlepac.adrizzle: num_cores : 1
INFO:drizzlepac.adrizzle: pixfrac : 1.0
INFO:drizzlepac.adrizzle: proc_unit : electrons
INFO:drizzlepac.adrizzle: rules_file : None
INFO:drizzlepac.adrizzle: stepsize : 10
INFO:drizzlepac.adrizzle: units : cps
INFO:drizzlepac.adrizzle: wcskey :
INFO:drizzlepac.adrizzle: wht_type : None
INFO:drizzlepac.adrizzle: wt_scl : exptime
INFO:drizzlepac.adrizzle: **Using sub-sampling value of 10 for kernel turbo
INFO:drizzlepac.adrizzle:Running Drizzle to create output frame with WCS of:
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 109.39116177250045 37.74673386265051
CRVAL : 109.39116177250045 37.74673386265051
INFO:astropy.wcs.wcs:CRPIX : 549.0 484.0
CRPIX : 549.0 484.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
INFO:astropy.wcs.wcs:NAXIS : 1098 968
NAXIS : 1098 968
INFO:drizzlepac.adrizzle:Executing serially
INFO:drizzlepac.adrizzle:-Drizzle input: ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.192738 to ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529rmq_single_sci.fits
-Generating simple FITS output: ica529rmq_single_sci.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rmq_single_sci.fits
Writing out image to disk: ica529rmq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rmq_single_wht.fits
Writing out image to disk: ica529rmq_single_wht.fits
INFO:drizzlepac.adrizzle:-Drizzle input: ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.000000 to ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529rsq_single_sci.fits
-Generating simple FITS output: ica529rsq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rsq_single_sci.fits
Writing out image to disk: ica529rsq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rsq_single_wht.fits
Writing out image to disk: ica529rsq_single_wht.fits
INFO:drizzlepac.adrizzle:-Drizzle input: ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.326927 to ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529s0q_single_sci.fits
-Generating simple FITS output: ica529s0q_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s0q_single_sci.fits
Writing out image to disk: ica529s0q_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s0q_single_wht.fits
Writing out image to disk: ica529s0q_single_wht.fits
INFO:drizzlepac.adrizzle:-Drizzle input: ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.029756 to ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529s6q_single_sci.fits
-Generating simple FITS output: ica529s6q_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s6q_single_sci.fits
Writing out image to disk: ica529s6q_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s6q_single_wht.fits
Writing out image to disk: ica529s6q_single_wht.fits
INFO:drizzlepac.util:==== Processing Step Separate Drizzle finished at 20:16:45.771 (02/12/2025)
==== Processing Step Separate Drizzle finished at 20:16:45.771 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Create Median started at 20:16:45.772 (02/12/2025)
==== Processing Step Create Median started at 20:16:45.772 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.createMedian:USER INPUT PARAMETERS for Create Median Step:
INFO:drizzlepac.createMedian: combine_bufsize : None
INFO:drizzlepac.createMedian: combine_grow : 1
INFO:drizzlepac.createMedian: combine_hthresh : None
INFO:drizzlepac.createMedian: combine_lthresh : None
INFO:drizzlepac.createMedian: combine_maskpt : 0.3
INFO:drizzlepac.createMedian: combine_nhigh : 0
INFO:drizzlepac.createMedian: combine_nlow : 0
INFO:drizzlepac.createMedian: combine_nsigma : 4 3
INFO:drizzlepac.createMedian: combine_type : median
INFO:drizzlepac.createMedian: compress : False
INFO:drizzlepac.createMedian: median : True
INFO:drizzlepac.createMedian: median_newmasks : True
INFO:drizzlepac.createMedian: proc_unit : native
INFO:drizzlepac.createMedian:reference sky value for image 'ica529rmq_flt.fits' is 29.476032680689162
reference sky value for image 'ica529rmq_flt.fits' is 29.476032680689162
INFO:drizzlepac.createMedian:reference sky value for image 'ica529rsq_flt.fits' is 0.0
reference sky value for image 'ica529rsq_flt.fits' is 0.0
INFO:drizzlepac.createMedian:reference sky value for image 'ica529s0q_flt.fits' is 49.99804987354285
reference sky value for image 'ica529s0q_flt.fits' is 49.99804987354285
INFO:drizzlepac.createMedian:reference sky value for image 'ica529s6q_flt.fits' is 6.038548978562271
reference sky value for image 'ica529s6q_flt.fits' is 6.038548978562271
INFO:drizzlepac.createMedian:Saving output median image to: 'ica529030_pcorr_med.fits'
Saving output median image to: 'ica529030_pcorr_med.fits'
INFO:drizzlepac.util:==== Processing Step Create Median finished at 20:16:45.934 (02/12/2025)
==== Processing Step Create Median finished at 20:16:45.934 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Blot started at 20:16:45.935 (02/12/2025)
==== Processing Step Blot started at 20:16:45.935 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.ablot:USER INPUT PARAMETERS for Blot Step:
INFO:drizzlepac.ablot: blot_addsky : True
INFO:drizzlepac.ablot: blot_interp : poly5
INFO:drizzlepac.ablot: blot_sinscl : 1.0
INFO:drizzlepac.ablot: blot_skyval : 0.0
INFO:drizzlepac.ablot: coeffs : True
INFO:drizzlepac.ablot: Blot: creating blotted image: ica529rmq_flt.fits[sci,1]
Blot: creating blotted image: ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.192738 to blotted image ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529rmq_sci1_blt.fits
-Generating simple FITS output: ica529rmq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rmq_sci1_blt.fits
Writing out image to disk: ica529rmq_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ica529rsq_flt.fits[sci,1]
Blot: creating blotted image: ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.000000 to blotted image ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529rsq_sci1_blt.fits
-Generating simple FITS output: ica529rsq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529rsq_sci1_blt.fits
Writing out image to disk: ica529rsq_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ica529s0q_flt.fits[sci,1]
Blot: creating blotted image: ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.326927 to blotted image ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529s0q_sci1_blt.fits
-Generating simple FITS output: ica529s0q_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s0q_sci1_blt.fits
Writing out image to disk: ica529s0q_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ica529s6q_flt.fits[sci,1]
Blot: creating blotted image: ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.029756 to blotted image ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ica529s6q_sci1_blt.fits
-Generating simple FITS output: ica529s6q_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ica529s6q_sci1_blt.fits
Writing out image to disk: ica529s6q_sci1_blt.fits
INFO:drizzlepac.util:==== Processing Step Blot finished at 20:16:46.707 (02/12/2025)
==== Processing Step Blot finished at 20:16:46.707 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Driz_CR started at 20:16:46.708 (02/12/2025)
==== Processing Step Driz_CR started at 20:16:46.708 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.drizCR:USER INPUT PARAMETERS for Driz_CR Step:
INFO:drizzlepac.drizCR: crbit : 4096
INFO:drizzlepac.drizCR: driz_cr : True
INFO:drizzlepac.drizCR: driz_cr_corr : False
INFO:drizzlepac.drizCR: driz_cr_ctegrow : 0
INFO:drizzlepac.drizCR: driz_cr_grow : 1
INFO:drizzlepac.drizCR: driz_cr_scale : 3.0 2.4
INFO:drizzlepac.drizCR: driz_cr_snr : 5.0 4.0
INFO:drizzlepac.drizCR: inmemory : False
INFO:drizzlepac.drizCR:Executing serially
INFO:drizzlepac.drizCR:Creating output: ica529rmq_sci1_crmask.fits
Creating output: ica529rmq_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ica529rsq_sci1_crmask.fits
Creating output: ica529rsq_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ica529s0q_sci1_crmask.fits
Creating output: ica529s0q_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ica529s6q_sci1_crmask.fits
Creating output: ica529s6q_sci1_crmask.fits
INFO:drizzlepac.util:==== Processing Step Driz_CR finished at 20:16:47.122 (02/12/2025)
==== Processing Step Driz_CR finished at 20:16:47.122 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Final Drizzle started at 20:16:47.124 (02/12/2025)
==== Processing Step Final Drizzle started at 20:16:47.124 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.adrizzle:Interpreted paramDict with single=False as:
{'build': True, 'stepsize': 10, 'coeffs': True, 'wcskey': '', 'wht_type': 'EXP', 'kernel': 'square', 'wt_scl': 'exptime', 'pixfrac': 1.0, 'fillval': None, 'maskval': None, 'bits': 528, 'units': 'cps'}
INFO:drizzlepac.adrizzle:USER INPUT PARAMETERS for Final Drizzle Step:
INFO:drizzlepac.adrizzle: bits : 528
INFO:drizzlepac.adrizzle: build : True
INFO:drizzlepac.adrizzle: clean : True
INFO:drizzlepac.adrizzle: coeffs : True
INFO:drizzlepac.adrizzle: crbit : 4096
INFO:drizzlepac.adrizzle: fillval : None
INFO:drizzlepac.adrizzle: kernel : square
INFO:drizzlepac.adrizzle: logfile : astrodrizzle.log
INFO:drizzlepac.adrizzle: maskval : None
INFO:drizzlepac.adrizzle: pixfrac : 1.0
INFO:drizzlepac.adrizzle: proc_unit : native
INFO:drizzlepac.adrizzle: rules_file : None
INFO:drizzlepac.adrizzle: stepsize : 10
INFO:drizzlepac.adrizzle: units : cps
INFO:drizzlepac.adrizzle: wcskey :
INFO:drizzlepac.adrizzle: wht_type : EXP
INFO:drizzlepac.adrizzle: wt_scl : exptime
INFO:drizzlepac.adrizzle: **Using sub-sampling value of 10 for kernel square
INFO:drizzlepac.adrizzle:Removing previous output product...
INFO:drizzlepac.adrizzle:Running Drizzle to create output frame with WCS of:
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 109.39116177250045 37.74673386265051
CRVAL : 109.39116177250045 37.74673386265051
INFO:astropy.wcs.wcs:CRPIX : 549.0 484.0
CRPIX : 549.0 484.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
CD1_1 CD1_2 : -2.9182452866968602e-05 -2.0433431583842394e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
CD2_1 CD2_2 : -2.0433431583842394e-05 2.9182452866968602e-05
INFO:astropy.wcs.wcs:NAXIS : 1098 968
NAXIS : 1098 968
INFO:drizzlepac.adrizzle:-Drizzle input: ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.192738 to ica529rmq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.000000 to ica529rsq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.326927 to ica529s0q_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.029756 to ica529s6q_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.outputimage:-Generating multi-extension output file: ica529030_pcorr_drz.fits
-Generating multi-extension output file: ica529030_pcorr_drz.fits
INFO:stwcs.wcsutil.altwcs:Deleted all instances of WCS with key A in extensions [1]
Deleted all instances of WCS with key A in extensions [1]
INFO:stwcs.wcsutil.altwcs:Deleted all instances of WCS with key B in extensions [1]
Deleted all instances of WCS with key B in extensions [1]
INFO:drizzlepac.outputimage:Writing out to disk: ica529030_pcorr_drz.fits
Writing out to disk: ica529030_pcorr_drz.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.util:==== Processing Step Final Drizzle finished at 20:16:48.515 (02/12/2025)
==== Processing Step Final Drizzle finished at 20:16:48.515 (02/12/2025)
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:AstroDrizzle Version 3.10.0 is finished processing at 20:16:48.516 (02/12/2025).
AstroDrizzle Version 3.10.0 is finished processing at 20:16:48.516 (02/12/2025).
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.util:
INFO:drizzlepac.util: -------------------- --------------------
-------------------- --------------------
INFO:drizzlepac.util: Step Elapsed time
Step Elapsed time
INFO:drizzlepac.util: -------------------- --------------------
-------------------- --------------------
INFO:drizzlepac.util:
INFO:drizzlepac.util: Initialization 0.5406 sec.
Initialization 0.5406 sec.
INFO:drizzlepac.util: Static Mask 0.0725 sec.
Static Mask 0.0725 sec.
INFO:drizzlepac.util: Subtract Sky 1.0882 sec.
Subtract Sky 1.0882 sec.
INFO:drizzlepac.util: Separate Drizzle 1.0569 sec.
Separate Drizzle 1.0569 sec.
INFO:drizzlepac.util: Create Median 0.1614 sec.
Create Median 0.1614 sec.
INFO:drizzlepac.util: Blot 0.7719 sec.
Blot 0.7719 sec.
INFO:drizzlepac.util: Driz_CR 0.4144 sec.
Driz_CR 0.4144 sec.
INFO:drizzlepac.util: Final Drizzle 1.3909 sec.
Final Drizzle 1.3909 sec.
INFO:drizzlepac.util: ==================== ====================
==================== ====================
INFO:drizzlepac.util: Total 5.4968 sec.
Total 5.4968 sec.
INFO:drizzlepac.util:
INFO:drizzlepac.imageObject:Removing intermediate files for ica529rmq_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ica529rsq_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ica529s0q_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ica529s6q_flt.fits
INFO:drizzlepac.util:Trailer file written to: astrodrizzle.log
Trailer file written to: astrodrizzle.log
3.4 Compare the original and corrected DRZ files #
Here, we display the drizzled image from the pipeline and the reprocessed drizzled image with persistence masked.
drz = fits.getdata('ica529030_drz.fits', ext=1)
drz_corr = fits.getdata('ica529030_pcorr_drz.fits', ext=1)
fig = plt.figure(figsize=(20, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(drz, vmin=0.85, vmax=1.4, cmap='Greys_r', origin='lower')
ax2.imshow(drz_corr, vmin=0.85, vmax=1.4, cmap='Greys_r', origin='lower')
ax1.set_title('ica529030_drz.fits (Pipeline SCI)', fontsize=20)
ax2.set_title('ica529030_pcorr_drz.fits (Corrected SCI)', fontsize=20)
ax1.plot([0, 280, 280, 0, 0], [250, 250, 350, 350, 250], c='red')
ax2.plot([0, 280, 280, 0, 0], [250, 250, 350, 350, 250], c='red')
ax1.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
ax2.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
[<matplotlib.lines.Line2D at 0x7f08edb06d90>]

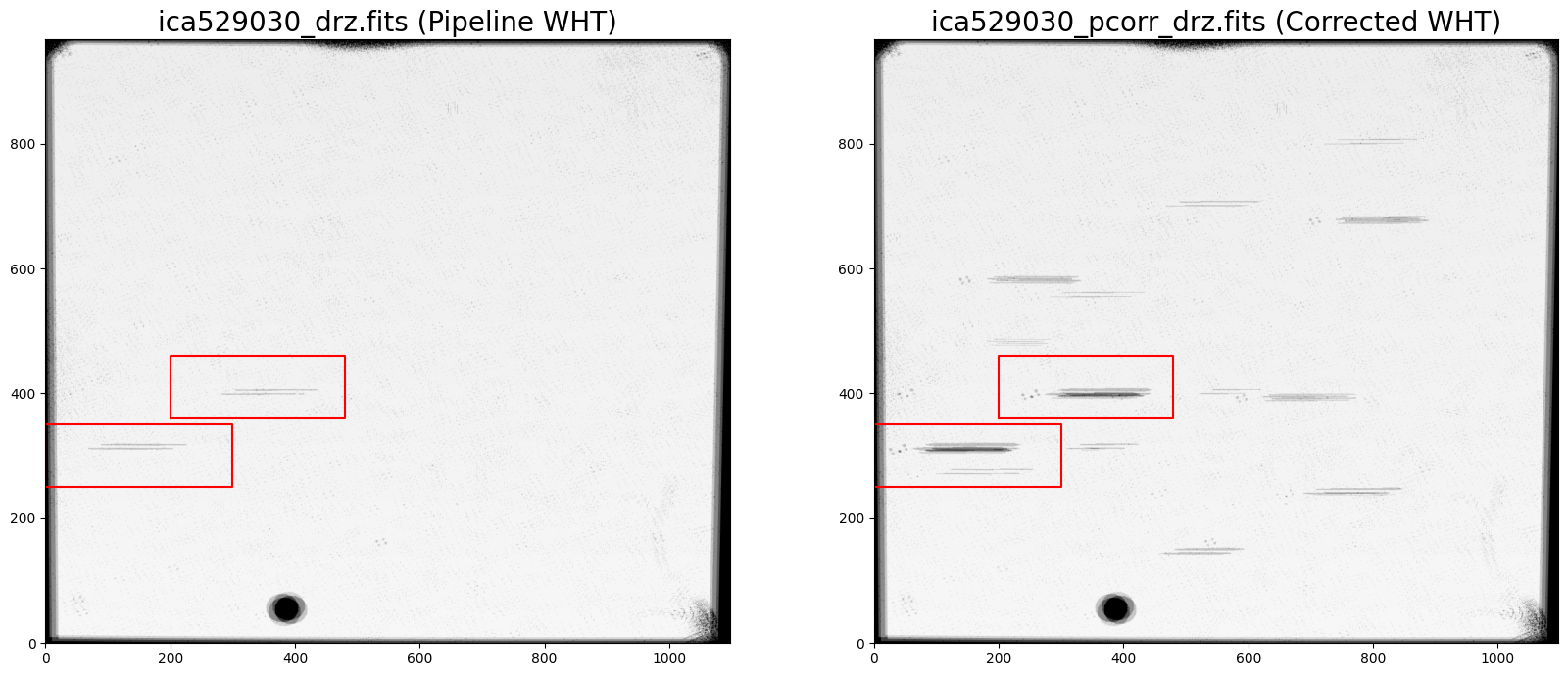
3.5 Compare the original and corrected WHT files #
When final_wht_type='EXP', the drizzled weight (WHT) images provide an effective exposure time map of the combined array.
In the plots below, we can see that the pipeline products have lower weight in the region impacted by the grism, but that only a single frame was flagged and rejected by the cosmic ray algorithm. The grey horizontal bars have a weight ~500 seconds compared to the total exposure ~700 seconds.
In the redrizzled ‘corrected’ WHT image, only a single exposure contributed to the darkest horizontal bars which have a value of ~200 seconds. The adjacent grey bars have a weight ~500 seconds and the rest of the WHT image is ~700 seconds. Users will need to experiment with the persistence masking thresholds and consider the size of the dithers in their individual datasets to determine the best masking strategy.
wht = fits.getdata('ica529030_drz.fits', ext=2)
wht_corr = fits.getdata('ica529030_pcorr_drz.fits', ext=2)
fig = plt.figure(figsize=(20, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(wht, vmin=0, vmax=800, cmap='Greys_r', origin='lower')
ax2.imshow(wht_corr, vmin=0, vmax=800, cmap='Greys_r', origin='lower')
ax1.set_title('ica529030_drz.fits (Pipeline WHT)', fontsize=20)
ax2.set_title('ica529030_pcorr_drz.fits (Corrected WHT)', fontsize=20)
ax1.plot([0, 300, 300, 0, 0], [250, 250, 350, 350, 250], c='red')
ax2.plot([0, 300, 300, 0, 0], [250, 250, 350, 350, 250], c='red')
ax1.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
ax2.plot([200, 480, 480, 200, 200], [360, 360, 460, 460, 360], c='red')
[<matplotlib.lines.Line2D at 0x7f08ed8f5550>]
4. Conclusions #
Thank you for walking through this notebook. Now with WFC3 data, you should be familiar with:
Examining the persistence models for a given dataset.
Defining a threshold to use for masking pixels in the DQ array of FLT science frames.
Reprocessing dithered frames with the new DQ flags to produce an improved combined DRZ image.
Congratulations, you have completed the notebook.
Additional Resources #
Below are some additional resources that may be helpful. Please send any questions through the HST Helpdesk.
-
see Sections 5.7.9 and 7.9.4 for a discussion of IR persistence
-
see Section 8.1 for a discussion of IR persistence
see Section 8.2 for evaluating the amount of persistence in science images
see Section 8.3 for a discussion methods for mitigating persistence
About this Notebook #
Author: Jennifer Mack, WFC3 Instrument Team
Created On: 2021-09-28
Updated On: 2023-11-06
Source: The notebook is sourced from hst_notebooks/notebooks/WFC3/persistence.
Citations #
If you use astropy, astroquery, ccdproc, or drizzlepac for published research, please cite the
authors. Follow these links for more information about citing the libraries below:


