
Processing WFC3/UVIS Data with calwf3 Using the v1.0 CTE-Correction#
Learning Goals#
This notebook explains how to calibrate raw WFC3/UVIS data with the v1.0 pixel-based CTE correction within calwf3.
By the end of this tutorial, you will:
Download a raw WFC3 image from MAST.
Find the necessary reference files needed for calibration.
Edit header keywords.
Run
calwf3v3.5.2to calibrate the raw image with the v1.0 pixel based CTE-correction.Compare v1.0 and v2.0 products.
Table of Contents#
1. Imports
2. Verify archived_drkcfiles.txt is in CWD
3. Check that calwf3 Version is v3.5.2
4. Query MAST and Download a WFC3 raw.fits Image
5. Set CRDS Environment Variable and Download Reference Files
5.1 Run crds bestrefs
5.2 Inspect Image Header
6. Find the Correct v1.0 DRKCFILE
6.1 Download the Correct DRKCFILE from CRDS
6.2 Modify the Image Header Keyword, DRKCFILE
7. Download the v1.0 PCTETAB
7.1 Modify the Image Header Keyword PCTETAB
8. Re-Inspect Image Header
9. Run calwf3
10. Inspect FLC Image Header
11. Investigate v1.0 and v2.0 Differences
11.1 Download the v2.0 FLC File
11.2 Open Files
11.3 Display 50x50 Pixel Background Subsection
11.3.1 Pixel Distribution of Background Subsections
11.4 Display Image Subsection
11.5 Aperture Photometry
12. Conclusions
Introduction#
The v1.0 pixel-based Charge Transfer Efficiency (CTE) correction was first implemented into calwf3 v3.3 in 2016
(Ryan et al. 2016, Anderson & Bedin 2010, HSTCAL release notes). This also marked the first time users could directly
download CTE-corrected flc & drc files from MAST. While the v1.0 correction was sufficient for many years, the
degradation of CTE over time reduced the efficacy of the model in treating low-level pixels. The v1.0 correction adversely
impacts (overcorrects) both the image background and faint sources. In April 2021 the v2.0 pixel-based CTE correction
was implemented in calwf3 v3.6.0 (Anderson et al. 2021, Kuhn & Anderson 2021, HSTCAL release notes). Since
MAST uses the latest release of calwf3 for calibration, any WFC3/UVIS CTE corrected data retrieved from MAST,
regardless of observation date, will be calibrated with the v2.0 pixel-based CTE correction. Although v1.0 pixel-based
CTE-corrected flc & drc files are no longer accessable through MAST, this notebook steps through the procedure
required to calibrate WFC3/UVIS images using the v1.0 CTE correction.
One of the limiting factors of using the v1.0 CTE correction are the CTE corrected dark current reference files (DRKCFILE).
These dark reference files are delivered to MAST by the WFC3 team and use the same pixel-based CTE correction within
calwf3. Now that we have switched to the v2.0 CTE correction there is a cut off for dark current reference files that use
the v1.0 correction. Observations taken after February 2021 will not have CTE corrected dark files using the v1.0 algorithm,
which means applying the v1.0 CTE correction works best for observations taken between May 2009 - February 2021.
If the observation being calibrated was taken after February 2021 there are two options: 1) use the last v1.0 CTE corrected
dark reference file from February 2021 or 2) use the v2.0 CTE corrected dark with the most appropriate USEAFTER for the
science exposure’s observation date.
1. Imports#
We import:
Package Name |
Purpose |
|---|---|
|
creating list of files |
|
directory maintenance and setting environment variables |
|
opening and modifying fits files |
|
downloading data from MAST |
|
creating and manipulating data tables |
|
finding z-scale limits when displaying images |
|
plotting and displaying images |
|
finding indices and concatenating arrays |
|
performing aperture photometry |
|
creating circular apertures |
|
creating circular annuli |
|
verifying the version and running pipeline |
|
measuring background values within annuli |
import glob
import os
from astropy.io import fits
from astropy.table import Table
from astroquery.mast import Observations
from astropy.visualization import ZScaleInterval
import matplotlib.pyplot as plt
import numpy as np
from photutils.aperture import aperture_photometry, CircularAperture, CircularAnnulus
from wfc3tools import calwf3
from example.background_median import aperture_stats_tbl
2. Verify archived_drkcfiles.txt is in CWD#
When you cloned/downloaded this notebook from hst_notebooks, a .txt file should have been included. The file name is
archived_drkcfiles.txt and it is used later on in the notebook. This .txt file includes the file name, delivery date, activation
date, and USEAFTER date for every v1.0 CTE corrected dark reference file between May 2009 - February 2021. Below, we will use
this file in conjunction with the observation date of the file(s) being calibrated to pick out the most appropriate v1.0 CTE corrected
dark reference file(s).
Please make sure the archived_drkcfiles.txt file is in the current working directory before continuing.
# list cwd to verify txt file is there
!ls -l archived_drkcfiles.txt
-rw-r--r-- 1 runner runner 350644 Dec 2 20:14 archived_drkcfiles.txt
3. Check that calwf3 Version is v3.5.2#
In April 2021, a new calwf3 version was released that contains the v2.0 CTE-correction. If you would like to use the v1.0 correction,
your current environment must be using calwf3 versions equal to or between 3.3 - 3.5.2. However, in order to get the best
v1.0 calibrated images we must use calwf3 v3.5.2. All calwf3 versions are hosted in the hstcal package, and calwf3
v3.5.2 is part of the hstcal release v2.6.0. This version of calwf3 also includes the (~Jan 2021) update that added MJD
as a parameterized variable for the PHOTMODE keyword, which enables a time-dependent photometric correction and zeropoint.
If your version of calwf3 is 3.6.0 or higher, you must downgrade the hstcal package. The safer option, however, is to
create a new environment using the requirements file provided in the notebook’s repository.
We recommend that you create an environment with the following commands in order to run this notebook properly:
$ conda create -n v1_PCTE -c conda-forge hstcal==2.6.0 python==3.11$ conda activate v1_PCTE$ pip install -r requirements.txt
hstcal v2.6.0 provides version 3.5.2 of calwf3, which is the last version that offers the v1.0 pixel-based CTE correction.
# print calwf3 version to make sure its equal to or between 3.3 and 3.5.2
!calwf3.e --version
3.7.2
4. Query MAST and Download a WFC3 raw.fits Image#
Here, we download our image via astroquery. For more information, please look at the documentation for
Astroquery,
Astroquery.mast, and
CAOM Field Descriptions, which is used for the obs_table variable below.
Additionally, you may download the data from MAST using either the HST MAST Search Engine or the more
general MAST Portal.
We download a raw image of star cluster 47 Tucanae (47Tuc, NGC 104), offset from the core, from CAL proposal 15576 (July 2019).
After downloading the image, we move it to the current working directory (cwd).
# Edit this cell's first line if you would to download your own file(s)
# Get the observation records
obs_table = Observations.query_criteria(obs_id='idv404axq*', proposal_id=15576)
# Get the listing of data products
products = Observations.get_product_list(obs_table)
# Filter the products for the RAW files
filtered_products = Observations.filter_products(products, productSubGroupDescription='RAW')
# Download all the images above
download_table = Observations.download_products(filtered_products, mrp_only=False)
# For convenience move raws to cwd and remove empty download dir
for file in download_table['Local Path']:
filename = file.split('/')[-1]
os.rename(file, os.path.basename(file))
os.rmdir('mastDownload/HST/'+filename[:9])
os.rmdir('mastDownload/HST/')
os.rmdir('mastDownload/')
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/idv404axq_raw.fits to ./mastDownload/HST/idv404axq/idv404axq_raw.fits ...
[Done]
# show list of current dir to verify fits file is there
!ls -l *raw.fits
-rw-r--r-- 1 runner runner 34891200 Dec 2 20:23 idv404axq_raw.fits
5. Set CRDS Environment Variable and Download Reference Files#
Before we run crds bestfefs and calwf3, we need to set environment variables for several subsequent calibration tasks.
We will point to a subdirectory within the main crds_cache/ using the IREF environment variable. The IREF variable is used
for WFC3 reference files. Other instruments use other variables, e.g., JREF for ACS. You have the option to permanently add these
environment variables to your user profile by adding the path in your shell’s configuration file. If you’re using bash, you would edit
the ~/.bash_profile file with lines such as:
export CRDS_SERVER_URL="https://hst-crds.stsci.edu"export CRDS_SERVER="https://hst-crds.stsci.edu"export CRDS_PATH="$HOME/crds_cache"export iref="${CRDS_PATH}/references/hst/wfc3/"
os.environ['CRDS_SERVER_URL'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_SERVER'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_PATH'] = 'crds_cache'
os.environ['iref'] = 'crds_cache/references/hst/wfc3/'
5.1 Run crds bestrefs#
The cell below calls CRDS bestref, which will copy the necessary reference files from CRDS over to your local machine, if you do not
already have them. Without running this command we would not be able to calibrate the image with calwf3.
!crds bestrefs --update-bestrefs --sync-references=1 --files idv404axq_raw.fits
CRDS - INFO - No comparison context or source comparison requested.
CRDS - INFO - ===> Processing idv404axq_raw.fits
CRDS - INFO - 0 errors
CRDS - INFO - 0 warnings
CRDS - INFO - 2 infos
5.2 Inspect Image Header#
When processing a raw file through calwf3 , the pipeline uses a few different header keywords to initiate and run the
pixel-based CTE correction. Here, we inspect the important header keywords from the raw file just downloaded.
At this step, you should see:
pctetabset toiref$54l1347ei_cte.fitsdrkcfileset toiref$54n2022fi_dkc.fitspctecorrset toPERFORM.
# Collect header keyword info from raw file
file, date, expstart, pctetab, drkcfile, pctecorr = [], [], [], [], [], []
for f in glob.glob('*raw.fits'):
h = fits.getheader(f)
file.append(h['filename'])
date.append(h['date-obs'])
expstart.append(h['expstart'])
pctetab.append(h['pctetab'])
drkcfile.append(h['drkcfile'])
pctecorr.append(h['pctecorr'])
image_table = Table([file, date, expstart, pctetab, drkcfile, pctecorr],
names=('file', 'date-obs', 'expstart', 'pctetab', 'drkcfile', 'pctecorr'))
image_table['expstart'].format = '5.6f'
# Sort and display the table
image_table.sort('expstart')
image_table
| file | date-obs | expstart | pctetab | drkcfile | pctecorr |
|---|---|---|---|---|---|
| str18 | str10 | float64 | str23 | str23 | str7 |
| idv404axq_raw.fits | 2019-07-29 | 58693.181643 | iref$54l1347ei_cte.fits | iref$7bg1930ki_dkc.fits | PERFORM |
6. Find the Correct v1.0 DRKCFILE#
Below, we open the .txt file containing a list of all the DRKCFILE reference files created with the v1.0 pixel-based CTE correction. DRKCFILE reference files are CTE corrected files used by the pipeline to perform the dark current subtraction during the generation
of the flc file. The DRKCFILE files listed in archived_drkcfiles.txt have been archived on the CRDS database and while
they are still accessible for use and download, they are not being actively used by MAST.
In the first cell, we generate an astropy.Table ( drkc_table ) using the data from the file archived_drkcfiles.txt,
mentioned in Section 2, and create empty lists for the final table. Then, in the second cell, we index the drkc_table table for the
best DRKCFILE that corresponds to the DATE-OBS of the raw file being calibrated. Lastly, in the third cell, we create and display
the final astropy.table that contains just the necessary DRKCFILE.
# Generate astropy table from `archived_drkcfiles.txt`
drkc_table = Table.read('archived_drkcfiles.txt', format='ascii.commented_header')
# Create empty lists for final astropy table
rawfiles, obsdates, dkcfiles, uafters, active_dates = [], [], [], [], []
# Using image header table from section 4.1 find closest drkcfile
for expstart in image_table['expstart']:
table_idx = np.where(abs(drkc_table['useafter-mjd']-expstart) == abs(drkc_table['useafter-mjd']-expstart).min())[0][0]
rawfile = image_table[image_table['expstart'] == expstart]['file'][0]
# if drkcfile has useafter date > rawfile expstart use previous drkcfile
if drkc_table[table_idx]['useafter-mjd'] > image_table[image_table['file'] == rawfile]['expstart'][0]:
table_idx -= 1
# append info
rawfiles.append(rawfile)
obsdates.append(image_table[image_table['file'] == rawfile]['date-obs'][0])
dkcfiles.append(drkc_table[table_idx]['drkcfile'])
uafters.append(drkc_table[table_idx]['useafter'])
active_dates.append(drkc_table[table_idx]['activation-date'])
# Generate table of filename, date-obs, drkc-filename, corresponding useafter
raw_dkc_tab = Table([rawfiles, obsdates, dkcfiles, uafters, active_dates],
names=('filename', 'date-obs', 'dkc-filename', 'dkc-useafter', 'dkc-activation'))
# Display table
raw_dkc_tab
| filename | date-obs | dkc-filename | dkc-useafter | dkc-activation |
|---|---|---|---|---|
| str18 | str10 | str18 | str10 | str10 |
| idv404axq_raw.fits | 2019-07-29 | 3961719li_dkc.fits | 2019-07-28 | 2019-09-09 |
6.1 Download the Correct DRKCFILE from CRDS#
Now that we know the name of the correct DRKCFILE, it must be retrieved from CRDS and stored on your local machine so that it
can be used during calibration. To copy the file from CRDS we use the crds sync command.
for dkc in raw_dkc_tab['dkc-filename']:
!crds sync --hst --files {dkc} --output-dir {os.environ['iref']}
CRDS - INFO - Symbolic context 'hst-latest' resolves to 'hst_1298.pmap'
CRDS - INFO - Reorganizing 27 references from 'flat' to 'flat'
CRDS - INFO - Syncing explicitly listed files.
CRDS - INFO - 0 errors
CRDS - INFO - 0 warnings
CRDS - INFO - 3 infos
6.2 Modify the Image Header Keyword, DRKCFILE#
With the v1.0 CTE-corrected dark current reference file that corresponds to our raw science file copied to our local machine,
we’re ready to edit the header keyword with the proper DRKCFILE.
for file in glob.glob('i*raw.fits'):
# Using raw_dkc_tab from above, grab appropriate drkcfile
ctecorr_dark = 'iref$'+raw_dkc_tab[raw_dkc_tab['filename'] == file]['dkc-filename'][0]
fits.setval(file, 'DRKCFILE', value=ctecorr_dark)
7. Download the v1.0 PCTETAB#
The next reference file we’re going to download from CRDS is the PCTETAB. This is the pixel-based correction reference table
and without it the algorithm will not work. In order to use the v1.0 pixel based correction, we must retrieve the v1.0 PCTETAB
and set the header keyword to the proper reference file. In the cells below, we use the crds sync command again and then
set the raw file’s PCTETAB to the v1.0 reference table, zcv2057mi_cte.fits.
!crds sync --hst --files zcv2057mi_cte.fits --output-dir {os.environ['iref']}
CRDS - INFO - Symbolic context 'hst-latest' resolves to 'hst_1298.pmap'
CRDS - INFO - Reorganizing 27 references from 'flat' to 'flat'
CRDS - INFO - Syncing explicitly listed files.
CRDS - INFO - 0 errors
CRDS - INFO - 0 warnings
CRDS - INFO - 3 infos
7.1 Modify the Image Header Keyword, PCTETAB#
fits.setval('idv404axq_raw.fits', 'PCTETAB', value='iref$zcv2057mi_cte.fits')
8. Re-Inspect Image Header#
Now with the headers modified, we inspect the keywords one last time to verify the file was updated properly before we process
it through calwf3. At this point you should see:
PCTETABset toiref$zcv2057mi_cte.fitsDRKCFILEset toiref$3961719li_dkc.fits
# Recollect and display header keywords
file, date, expstart, pctetab, drkcfile, pctecorr = [], [], [], [], [], []
for f in glob.glob('*raw.fits'):
h = fits.getheader(f)
file.append(h['filename'])
date.append(h['date-obs'])
expstart.append(h['expstart'])
pctetab.append(h['pctetab'])
drkcfile.append(h['drkcfile'])
pctecorr.append(h['pctecorr'])
updated_table = Table([file, date, expstart, pctetab, drkcfile, pctecorr],
names=('file', 'date-obs', 'expstart', 'pctetab', 'drkcfile', 'pctecorr'))
updated_table['expstart'].format = '5.6f'
# Sort and display the table
updated_table.sort('expstart')
updated_table
| file | date-obs | expstart | pctetab | drkcfile | pctecorr |
|---|---|---|---|---|---|
| str18 | str10 | float64 | str23 | str23 | str7 |
| idv404axq_raw.fits | 2019-07-29 | 58693.181643 | iref$zcv2057mi_cte.fits | iref$3961719li_dkc.fits | PERFORM |
9. Run calwf3#
As a reminder, the calwf3 version must be 3.5.2 to use the v1.0 pixel-base CTE correction with the most up-to-date
calibration parameters. If you are not using version 3.5.2 the below calwf3 call will crash due to an out-of-date
IMPHTTAB reference file.
if not os.path.exists('idv404axq_flt.fits'):
calwf3('idv404axq_raw.fits')
# show list of cwd to verify calibrated files were made
!ls -ltr *.fits
-rw-r--r-- 1 runner runner 168079680 Dec 2 20:22 idv404axq_flt.fits
-rw-r--r-- 1 runner runner 168079680 Dec 2 20:22 idv404axq_flc.fits
-rw-r--r-- 1 runner runner 168802560 Dec 2 20:22 idv404axq_v2.0_flc.fits
-rw-r--r-- 1 runner runner 34891200 Dec 2 20:23 idv404axq_raw.fits
10. Inspect FLC Image Header#
To verify that the data was calibrated with the v1.0 pixel-based CTE-correction, header keyword CAL_VER should be 3.5.2,
CTE_VER should be 1.0, and CTE_NAME should be pixelCTE 2012
# Recollect and display FLC header keywords
file, pctetab, drkcfile, pctecorr, calver, ctename, ctever = [], [], [], [], [], [], []
for f in glob.glob('*flc.fits'):
h = fits.getheader(f)
file.append(h['filename'])
pctetab.append(h['pctetab'])
drkcfile.append(h['drkcfile'])
pctecorr.append(h['pctecorr'])
calver.append(h['cal_ver'])
ctename.append(h['cte_name'])
ctever.append(h['cte_ver'])
final_table = Table([file, pctetab, drkcfile, pctecorr, calver, ctename, ctever],
names=('file', 'pctetab', 'drkcfile', 'pctecorr', 'cal_ver', 'cte_name', 'cte_ver'))
final_table
| file | pctetab | drkcfile | pctecorr | cal_ver | cte_name | cte_ver |
|---|---|---|---|---|---|---|
| str18 | str23 | str23 | str8 | str19 | str13 | str3 |
| idv404axq_flc.fits | iref$zcv2057mi_cte.fits | iref$3961719li_dkc.fits | COMPLETE | 3.7.2 (Apr-15-2024) | pixelCTE 2012 | 1.0 |
| idv404axq_flc.fits | iref$54l1347ei_cte.fits | iref$7bg1930ki_dkc.fits | COMPLETE | 3.7.2 (Apr-15-2024) | pixelCTE 2019 | 2.0 |
11. Investigate v1.0 and v2.0 Differences#
Now that we have created a v1.0 CTE corrected image, lets compare it to the same image calibrated with the v2.0 correction.
11.1 Download the v2.0 FLC File#
First, we need to download the same FLC file from MAST that is corrected with the v2.0 pixel-based CTE correction so that we can
compare it to the v1.0 FLC file we just created in the notebook. In this step we rename the downloaded FLC to idv404axq_v2.0_flc.fits.
# Get the observation records
obs_table = Observations.query_criteria(obs_id='idv404axq*', proposal_id=15576)
# Get the listing of data products
products = Observations.get_product_list(obs_table)
# Filter the products for the RAW files
filtered_products = Observations.filter_products(products, productSubGroupDescription='FLC', project='CALWF3')
# Download all the images above
download_table = Observations.download_products(filtered_products, mrp_only=False)
# For convenience move raws to cwd and remove empty download dir
for file in download_table['Local Path']:
filename = file.split('/')[-1][:9]+'_v2.0_flc.fits'
os.rename(file, os.path.basename(filename))
os.rmdir('mastDownload/HST/'+filename[:9])
os.rmdir('mastDownload/HST/')
os.rmdir('mastDownload/')
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/idv404axq_flc.fits to ./mastDownload/HST/idv404axq/idv404axq_flc.fits ...
[Done]
11.2 Open Files#
In the cell below, we open each of the different files (FLT, v1.0 and v2.0 FLC) and create full-frame ~4Kx4K arrays. Additionally,
we multiply the science arrays by the pixel area maps. Due to the geometric distortion present in WFC3 images, these pixel
area maps are necessary to achieve uniformity in the measured counts of an object across the field. The pixel map simply
reflects the area of the pixels at the location of the source, and by multiplying the images by this field-dependent correction
factor, we will improve the accuracy of the photometry.
# Open data and set variables
with fits.open('idv404axq_v2.0_flc.fits') as hdu:
v2_uvis1 = hdu[4].data
v2_uvis2 = hdu[1].data
with fits.open('idv404axq_flc.fits') as hdu:
v1_uvis1 = hdu[4].data
v1_uvis2 = hdu[1].data
with fits.open('idv404axq_flt.fits') as hdu:
flt_uvis1 = hdu[4].data
flt_uvis2 = hdu[1].data
# Load pixel area maps
PAM_uvis1 = fits.getdata('example/UVIS1wfc3_map.fits')
PAM_uvis2 = fits.getdata('example/UVIS2wfc3_map.fits')
# Stich UVIS1 and 2 together and multiply by pixel area map
v2sci = np.concatenate([v2_uvis2*PAM_uvis2, v2_uvis1*PAM_uvis1])
v1sci = np.concatenate([v1_uvis2*PAM_uvis2, v1_uvis1*PAM_uvis1])
fltsci = np.concatenate([flt_uvis2*PAM_uvis2, flt_uvis1*PAM_uvis1])
11.3 Display 50x50 Pixel Background Subsection#
One of the differences between the v1.0 and v2.0 CTE correction is the background level. Here, we show a square 50x50 pixel
subsection that is mostly featureless, i.e. no known sources or cosmic ray hits. The v2.0 correction significantly reduces noise
amplification and improves the resulting background. To aid in the visual inspection of the background subsection, an animated
GIF is included in the notebook that blinks between the v1.0 and v2.0 FLC files.
# Generate subplots
fig, [ax1, ax2, ax3] = plt.subplots(1, 3, figsize=(15, 10), dpi=150)
# Generate background subsections
flt_bkg = fltsci[2070:2120, 2180:2230]
v1_bkg = v1sci[2070:2120, 2180:2230]
v2_bkg = v2sci[2070:2120, 2180:2230]
# Calculate min and max values for image scaling
z = ZScaleInterval()
z1, z2 = z.get_limits(v1_bkg)
# Display background subsection
im1 = ax1.imshow(flt_bkg, origin='lower', cmap='Greys_r', vmin=z1, vmax=z2)
im2 = ax2.imshow(v1_bkg, origin='lower', cmap='Greys_r', vmin=z1, vmax=z2)
im3 = ax3.imshow(v2_bkg, origin='lower', cmap='Greys_r', vmin=z1, vmax=z2)
# Formatting
fig.colorbar(im1, ax=ax1, shrink=0.35, pad=0.02)
fig.colorbar(im2, ax=ax2, shrink=0.35, pad=0.02)
fig.colorbar(im3, ax=ax3, shrink=0.35, pad=0.02)
ax1.set_title('FLT File BKG Subsection', size=14)
ax2.set_title('v1.0 PCTE FLC BKG Subsection', size=14)
ax3.set_title('v2.0 PCTE FLC BKG Subsection', size=14)
ax1.axis('off'), ax2.axis('off'), ax3.axis('off')
fig.tight_layout()
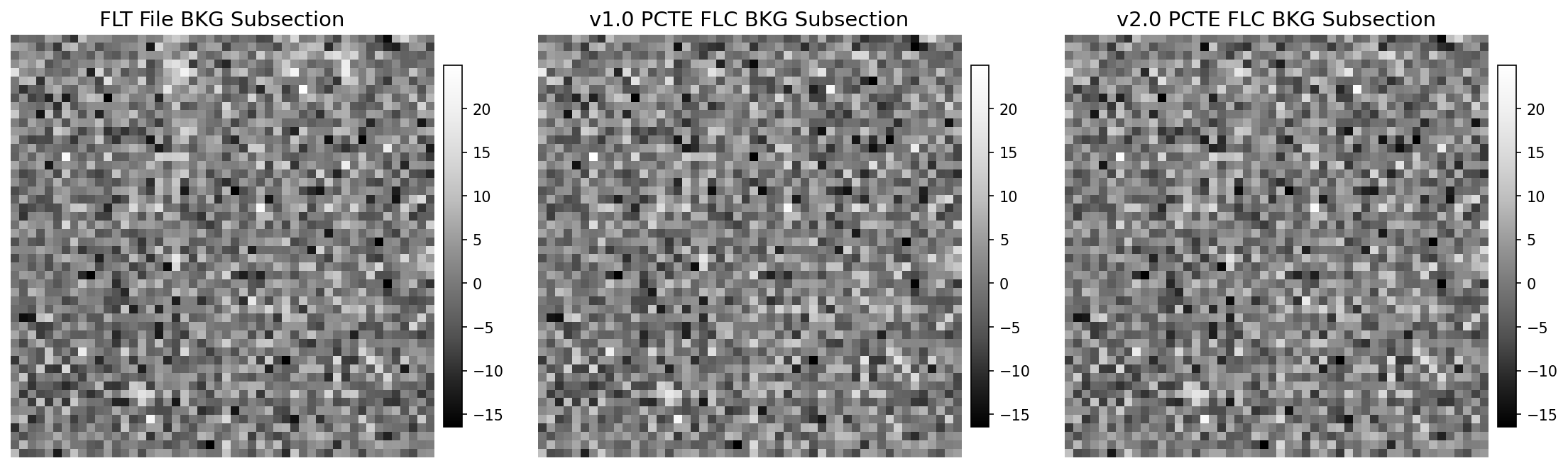
11.3.1 Pixel Distribution of Background Subsections#
To gain a more quantitative picture of how the background pixels are changing between the different file versions, we plot
the distribution of pixel values from the 50x50 pixel subsection above. The increased noise level in the v1.0 correction is
apparent in the blue histogram below. In addition, we have also computed the difference between the file types and have
plotted them as histograms. These two plots illustrates how the background in the v2.0 CTE corrected image is less noisy
and more in-line with the values seen in the FLT image.
# Generate subplots
fig, [ax1, ax2] = plt.subplots(2, 1, figsize=(7, 10), dpi=120)
# Plot background subsection histograms
ax1.hist(flt_bkg.ravel(), bins=100, range=(-30, 100), histtype='step', color='C3', label='FLT')
ax1.hist(v1_bkg.ravel(), bins=100, range=(-30, 100), histtype='step', color='C0', label='v1.0 FLC')
ax1.hist(v2_bkg.ravel(), bins=100, range=(-30, 100), histtype='step', color='k', label='v2.0 FLC')
# Plot background subsection differential histograms
ax2.hist((v1_bkg-flt_bkg).ravel(), bins=100, range=(-25, 50), histtype='step', color='magenta', label='v1.0 FLC $-$ FLT')
ax2.hist((v2_bkg-flt_bkg).ravel(), bins=100, range=(-25, 50), histtype='step', color='limegreen', label='v2.0 FLC $-$ FLT')
ax2.hist((v1_bkg-v2_bkg).ravel(), bins=100, range=(-25, 50), histtype='step', color='C9', label='v1.0 $-$ v2.0 FLC')
# Formatting
ax1.set_title('Background Subsection Histogram', size=14)
ax2.set_title('Background Subsection Differential Histogram', size=14)
ax1.set_xlabel('Pixel Value [e-]', size=12)
ax1.set_ylabel('Frequency', size=12)
ax2.set_xlabel('Pixel Value [e-]', size=12)
ax2.set_ylabel('Frequency', size=12)
ax1.grid(alpha=0.5), ax2.grid(alpha=0.5)
ax1.legend(), ax2.legend()
ax1.set_yscale('log')
ax2.set_yscale('log')
fig.tight_layout()
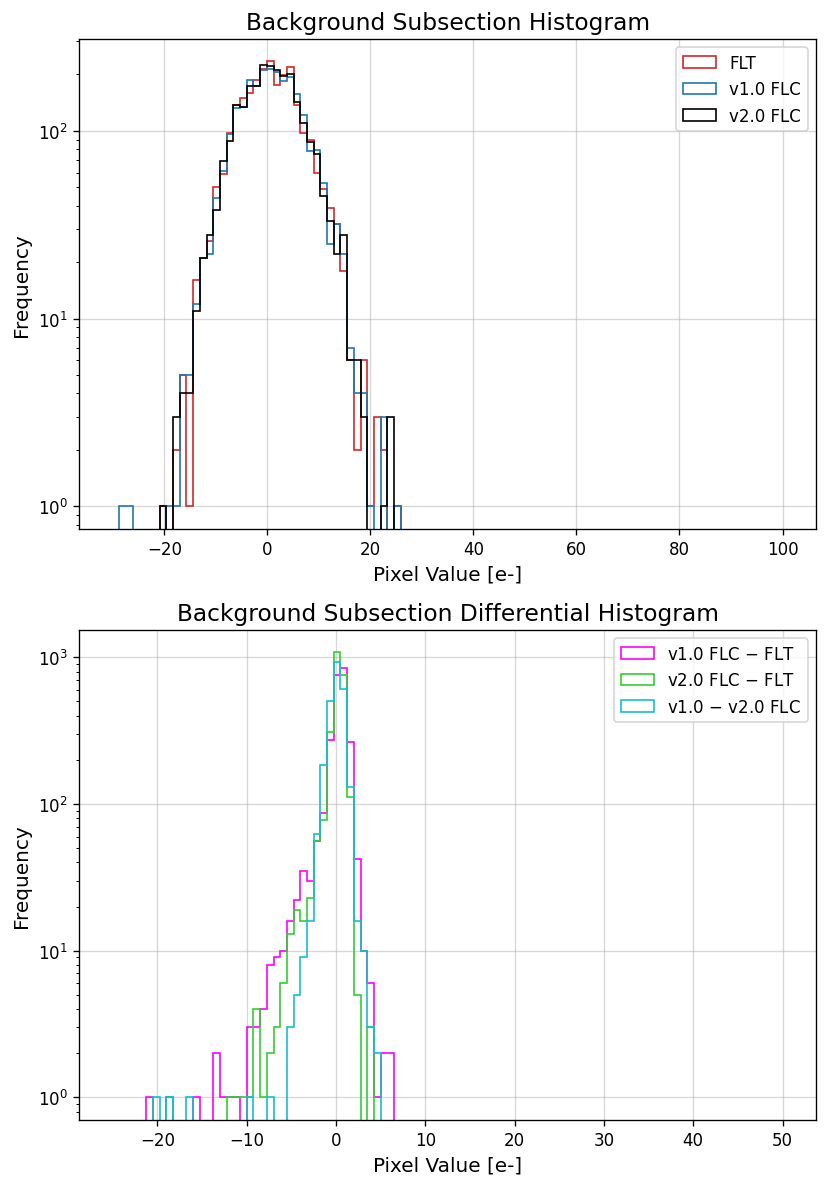
11.4 Display Image Subsection#
Another difference between the v1.0 and v2.0 pixel-based CTE model is the amount of correction applied to fainter sources.
In order to reduce noise amplification, the fluxes of faint sources only receive a limited amount of correction in the v2.0 version.
We again provide an animated GIF of the two subsections to aid with the visual comparison.
# Generate subplots
fig, [ax1, ax2] = plt.subplots(2, 1, figsize=(8, 8), dpi=150)
# Calculate min and max values for image scaling
z = ZScaleInterval()
z1, z2 = z.get_limits(v1_uvis1)
# Display subsection
im1 = ax1.imshow(v1_uvis1, origin='lower', cmap='Greys_r', vmin=z1, vmax=z2)
im2 = ax2.imshow(v2_uvis1, origin='lower', cmap='Greys_r', vmin=z1, vmax=z2)
# Formatting
ax1.set_xlim(85, 325), ax2.set_xlim(85, 325)
ax1.set_ylim(0, 149), ax2.set_ylim(0, 149)
fig.colorbar(im1, ax=ax1, shrink=0.95, pad=0.01)
fig.colorbar(im2, ax=ax2, shrink=0.95, pad=0.01)
ax1.set_title('v1.0 PCTE FLC UVIS1 Subsection', size=14)
ax2.set_title('v2.0 PCTE FLC UVIS1 Subsection', size=14)
fig.tight_layout()
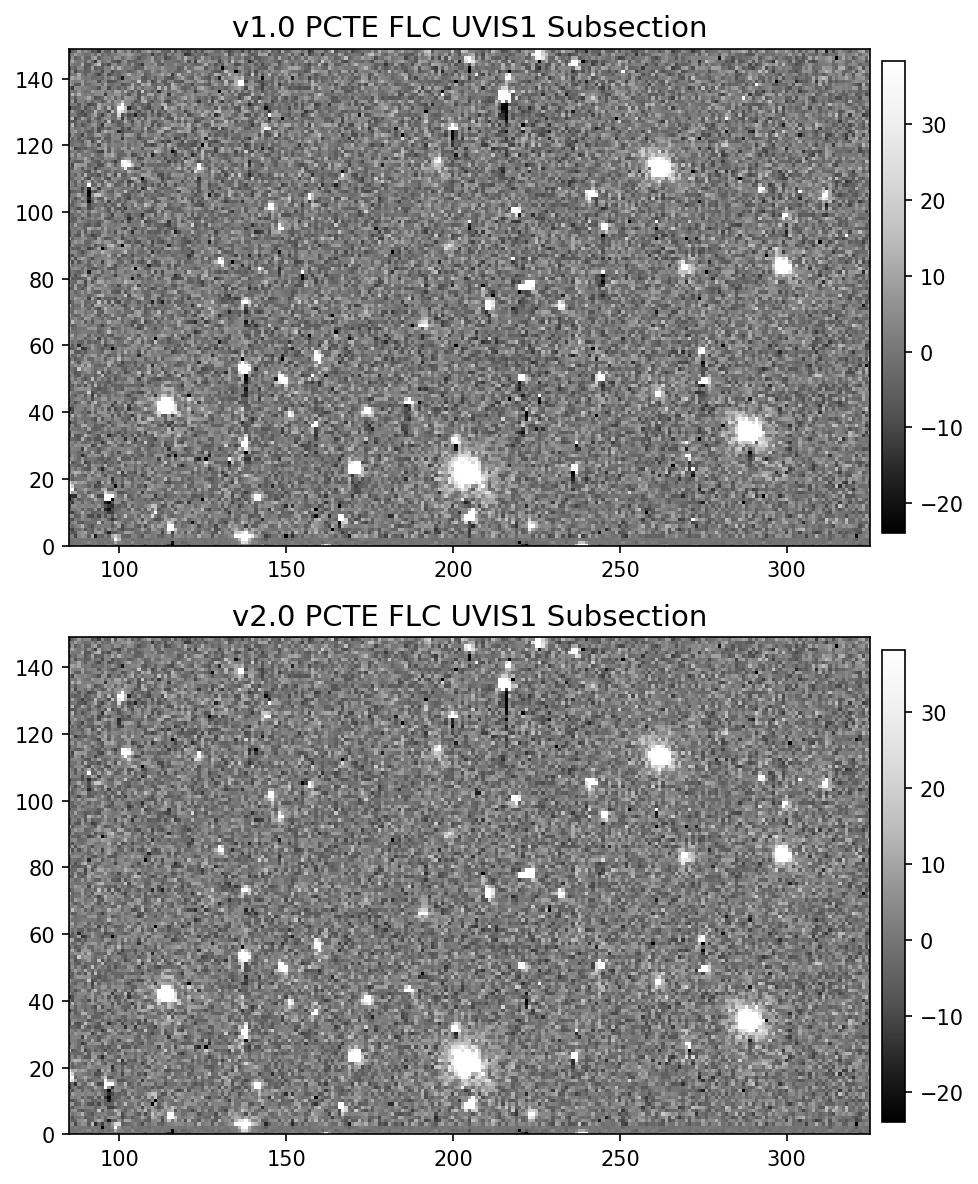
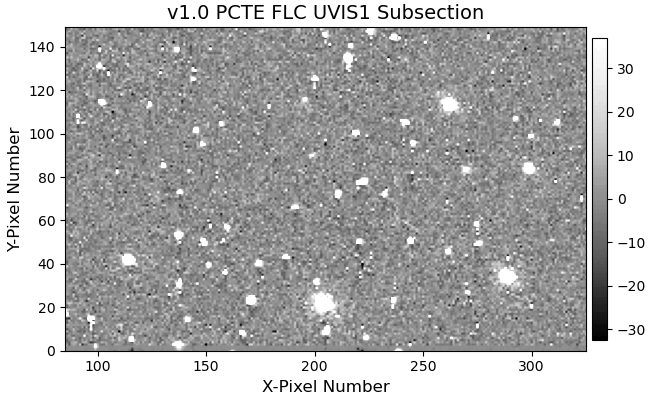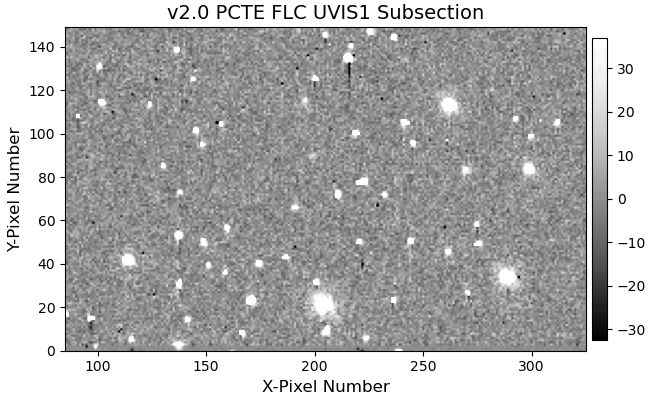
11.5 Aperture Photometry#
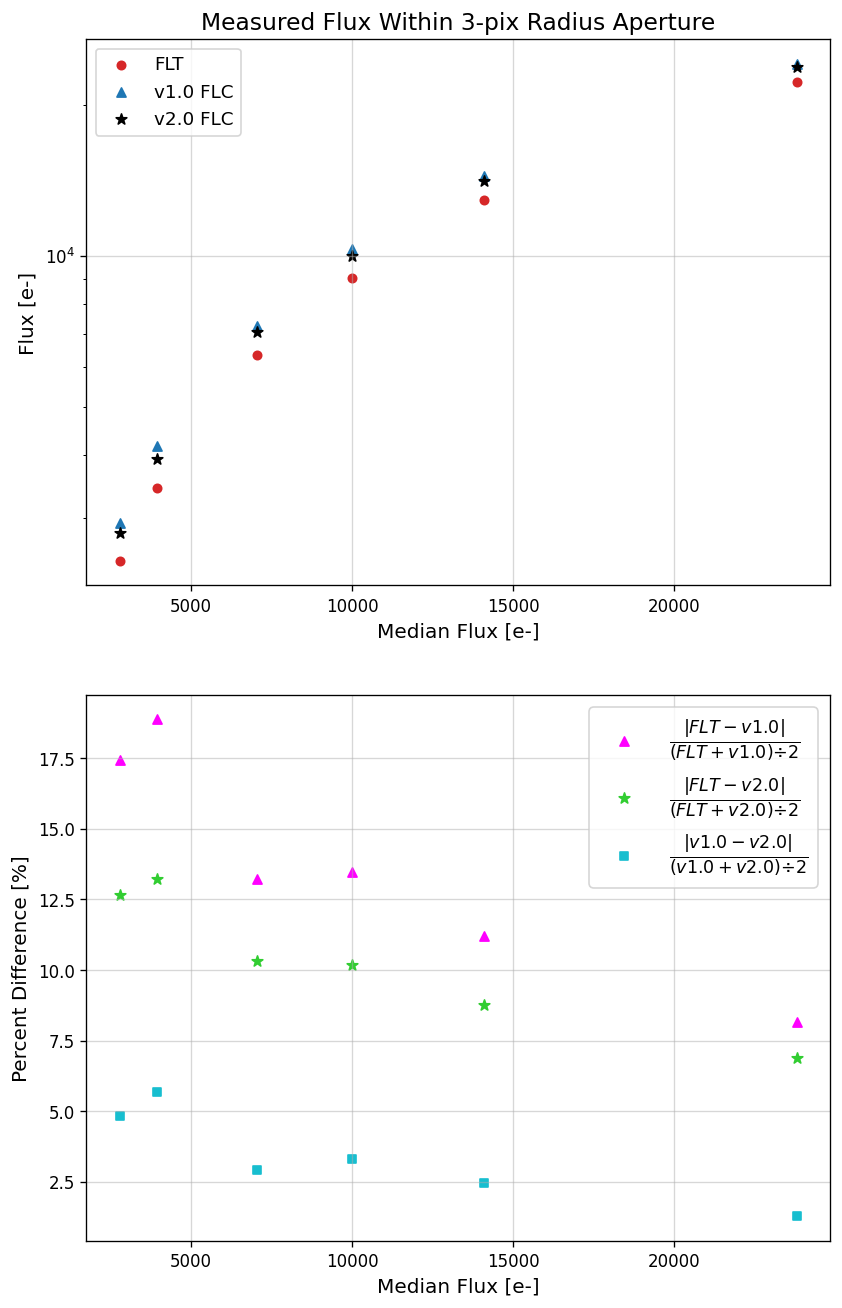
To show the quantitative difference in the observed flux of sources between the v1.0 and v2.0 pixel-based CTE correction,
we perform aperture photometry on six stars of varying brightness within the image subsection above. Here, we display the
image subsection again with the apertures, annuli, and star labels overplotted. This subsection includes the first ~150 rows
of UVIS 1, which means all of these stars suffer the most CTE flux loss as they transfer 1900+ rows to the readout amplifier.

First, we approximate the center x and y positions of the stars in the 4Kx4K science arrays, and create the apertures and
annuli using photutils.
Then, we use photutils to measure the signal in each area and subtract the background values in the annuli from the
aperture sum values. We define a function named get_flux() to do this for us.
def get_flux(data, aperture, annulus_aperture):
"""
Function to calculate background subtracted aperture sum
Parameters:
-----------
data : float array
The 2d array of science pixels being measured
aperture : photutils obj
A photutils aperture object with defined position and radius
annulus_aperture : photutils obj
A photutils circular annulus aperture object with defined position and radii
Return:
-------
flux : float
The measured background subtracted aperture sum
"""
# Generate photutils.aperture_photometry table object
phot = aperture_photometry(data, aperture)
# Measure background around sources. aperture_stats_tbl() comes from background_median.py
bkg_phot = aperture_stats_tbl(data, annulus_aperture, method='exact', sigma_clip=True)
# Calculate background subtracted aperture sum
flux = phot['aperture_sum'] - bkg_phot['aperture_median'] * aperture.area
return flux
# Approximate x,y pixel locations of each star in the 4Kx4K array
positions = [(299.4, 2135.6),
(114.7, 2093.4),
(171.3, 2074.9),
(262.6, 2164.9),
(289.1, 2085.6),
(204.8, 2073.1)]
# Photutils cirular aperture object with small radius
aperture = CircularAperture(positions, r=3)
# Photutils circular annulus aperture object
annulus_aperture = CircularAnnulus(positions, r_in=13, r_out=19)
# Call function to calculate flux of stars
# THE RUNTIME WARNING MAY BE IGNORED
fltflux = get_flux(fltsci, aperture, annulus_aperture)
v1flux = get_flux(v1sci, aperture, annulus_aperture)
v2flux = get_flux(v2sci, aperture, annulus_aperture)
/home/runner/work/hst_notebooks/hst_notebooks/notebooks/WFC3/calwf3_v1.0_cte/example/background_median.py:88: RuntimeWarning: invalid value encountered in divide
values = cutout * mask.data / mask.data
/home/runner/work/hst_notebooks/hst_notebooks/notebooks/WFC3/calwf3_v1.0_cte/example/background_median.py:88: RuntimeWarning: invalid value encountered in divide
values = cutout * mask.data / mask.data
/home/runner/work/hst_notebooks/hst_notebooks/notebooks/WFC3/calwf3_v1.0_cte/example/background_median.py:88: RuntimeWarning: invalid value encountered in divide
values = cutout * mask.data / mask.data
RuntimeWarning: invalid value encountered in true_divide message. Finally, we plot our results.
The first plot below shows the measured flux of the six stars in each of the different calibrated products. Stars 1-6 are organized
by increasing flux, with Star 1 being the faintest and Star 6 being the brightest. The x-axis values correspond to the median flux
value measured in each of the three different files (FLT, v1.0, and v2.0) per star. The y-axis values show the individual flux values
for each star where the different colors and symbols represent the three file types. For example, the individual flux values (y-axis)
of Star 1 for the FLT, v1.0, and v2.0 files are ~2467, 3056, and 2799 e- respectively. The median of the three values is ~2799 e-,
which is the value plotted on the x-axis. The second plot illustrates the percent differences between the three different
flux values per star, and uses the same x-axis values as the first plot.
The FLT files have no CTE correction applied and thus have the lowest measured flux for each star due to the CTE flux loss.
Stars 1-4 have noticably different flux values between the v1.0 and v2.0 CTE corrections. In v1.0, the CTE algorithm is actually
over-correcting these fainter sources resulting in higher aperture photometry measurements. The flux values for stars 1-4 range
from ~3000 - 10000 e- within a 3-pixel radius aperture. The flux values between the v1.0 and v2.0 CTE corrections for stars
1-4 have percent differences of ~9 - 4%. Sources with more than ~10000 e- within a 3-pixel aperture (stars 5 & 6) have more
compareable flux values between the v1.0 and v2.0 CTE corrections, and have percent differences less than 2%.
# Generate subplots
fig, [ax1, ax2] = plt.subplots(2, 1, figsize=(8, 13), dpi=120)
ax1.grid(alpha=0.5), ax2.grid(alpha=0.5)
# Find median flux values between products
medflux = np.median([fltflux, v1flux, v2flux], axis=0)
# Scatter plot of measured flux
ax1.scatter(medflux, fltflux, 25, marker='o', c='C3', label='FLT')
ax1.scatter(medflux, v1flux, 30, marker='^', c='C0', label='v1.0 FLC')
ax1.scatter(medflux, v2flux, 45, marker='*', c='k', label='v2.0 FLC')
# Scatter plot of percentage difference
ax2.scatter(medflux, abs((fltflux-v1flux))/((fltflux+v1flux)/2)*100, 30,
marker='^', c='magenta', label=r'$\frac{|FLT - v1.0|}{(FLT + v1.0) ÷ 2}$')
ax2.scatter(medflux, abs((fltflux-v2flux))/((fltflux+v2flux)/2)*100, 45,
marker='*', c='limegreen', label=r'$\frac{|FLT - v2.0|}{(FLT + v2.0) ÷ 2}$')
ax2.scatter(medflux, abs((v1flux-v2flux))/((v1flux+v2flux)/2)*100, 25,
marker='s', c='C9', label=r'$\frac{|v1.0 - v2.0|}{(v1.0 + v2.0) ÷ 2}$')
# Formatting
ax1.set_title('Measured Flux Within 3-pix Radius Aperture', size=14)
ax1.set_xlabel('Median Flux [e-]', size=12)
ax1.set_ylabel('Flux [e-]', size=12)
ax2.set_xlabel('Median Flux [e-]', size=12)
ax2.set_ylabel('Percent Difference [%]', size=12)
ax1.legend(prop={'size': 11}), ax2.legend(prop={'size': 15})
ax1.set_yscale('log')
12. Conclusions#
Thank you for walking through this notebook. You now have everything you need to process your own files with the v1.0 CTE
correction within calwf3. Since completing this notebook you should be more familiar with:
Applying the v1.0 pixel-based CTE correction to 2009-2021 UVIS data.
Which header keywords of the
rawfits file to edit in order to use the v1.0 correction.How to find and download the appropriate
PCTETABandDRKCFILEreference files.Verifying your version of
calwf3and calibrating arawfile with the v1.0 CTE correction.Investigating the differences between v1.0 and v2.0 CTE corrected files.
Congratulations, you have completed the notebook!#
Additional Resources#
Below are some additional resources that may be helpful. Please feel free to contact the WFC3 Helpdesk for any questions.
About this Notebook#
Author: Benjamin Kuhn; WFC3 Instrument Team
Created: February 22, 2022
Last updated on: October 30, 2025
Citations#
If you use astropy, astroquery, numpy, matplotlib, photutils, or scipy for published research, please cite the authors.
Follow these links for more information about citing the libraries below:


