
Viewing STIS Data#
Table of Contents#
By the end of this tutorial, you will go through:
0. Introduction#
The Space Telescope Imaging Spectrograph (STIS) is a versatile imaging spectrograph installed on the Hubble Space Telescope (HST), covering a wide range of wavelengths from the near-infrared region into the ultraviolet.
This tutorial aims to prepare new users to begin analyzing STIS data by going through downloading data, reading and viewing spectra, and viewing STIS images.
There are three detectors on STIS: FUV-MAMA, NUV-MAMA, and CCD. While the detectors are designed for different scientific purposes and operate at a different wavelength, their data are organized in the same structure. Thus we are using FUV-MAMA data as an example in this notebook.
For a detailed overview of the STIS instrument, see the STIS Instrument Handbook.
For more information on STIS data analysis and operations, see the STIS Data Handbook.
Defining some terms:
HST: Hubble Space Telescope
STIS: Space Telescope Imaging Spectrograph on HST (https://www.stsci.edu/hst/instrumentation/stis)
STIS/NUV-MAMA: Cs2Te Multi-Anode Microchannel Array (MAMA) detector for observing mainly in the near ultraviolet (NUV)
STIS/FUV-MAMA: Solar-blind CsI Multi-Anode Microchannel Array (MAMA) detector for observing mainly in the far ultraviolet (FUV)
CCD: Charge Coupled Device
FITS: Flexible Image Transport System (https://fits.gsfc.nasa.gov/fits_primer.html)
HDU: Header/Data Unit in a FITS file
0.1 Import necessary packages#
We will import the following packages:
astropy.io.fitsfor accessing FITS filesastropy.table.Tablefor creating tidy tables of the dataastroquery.mast.Observationsfor finding and downloading data from the MAST archivepathlibfor managing system pathsmatplotlib.pyplotfor plotting dataIPython.displayfor formatting displaynumpyto handle array functionspandasto make basic tables and dataframesstistoolsfor operations on STIS data
# Import for: Reading in fits file
from astropy.table import Table
from astropy.io import fits
# Import for: Downloading necessary files. (Not necessary if you choose to collect data from MAST)
from astroquery.mast import Observations
# Import for: Managing system variables and paths
from pathlib import Path
# Import for: Plotting and specifying plotting parameters
import matplotlib.pyplot as plt
import matplotlib
from IPython.display import display
# Import for: Quick Calculation and Data Analysis
import numpy as np
import pandas as pd
# Import for operations on STIS Data
import stistools
The following tasks in the stistools package can be run with TEAL:
basic2d calstis ocrreject wavecal x1d x2d
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/nmpfit.py:8: UserWarning: NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.
warnings.warn("NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.")
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/gfit.py:18: UserWarning: GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it.Use astropy.modeling instead.
warnings.warn("GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it."
1. Downloading STIS data from MAST using astroquery#
There are other ways to download data from MAST such as using CyberDuck. We are only showing how to use astroquery in this notebook
# make directory for downloading data
datadir = Path('./data')
datadir.mkdir(exist_ok=True)
%%capture --no-stderr --no-display
# Search target object by obs_id
target = Observations.query_criteria(obs_id='ODJ102010')
# get a list of files assiciated with that target
FUV_list = Observations.get_product_list(target)
# Download only the SCIENCE fits files
downloads = Observations.download_products(
FUV_list,
productType='SCIENCE',
extension='fits',
download_dir=str(datadir))
downloads
| Local Path | Status | Message | URL |
|---|---|---|---|
| str103 | str8 | object | object |
| data/mastDownload/HST/odj102010/odj102010_raw.fits | COMPLETE | None | None |
| data/mastDownload/HST/odj102010/odj102010_x2d.fits | COMPLETE | None | None |
| data/mastDownload/HST/odj102010/odj102010_flt.fits | COMPLETE | None | None |
| data/mastDownload/HST/odj102010/odj102010_x1d.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15168_stis_-alf-cep_odj1/hst_15168_stis_-alf-cep_g140m_odj1_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15168_stis_-alf-cep_odj1/hst_15168_stis_-alf-cep_g140m_odj102_cspec.fits | COMPLETE | None | None |
2. Reading in the data#
2.1 Investigating the data - Basics#
Before doing any operations on the data, we want to first explore the basics and data file structures.
The 1D-extracted, background subtracted, flux and wavelength calibrated spectra data are stored in a FITS file with suffix _x1d (note that for the CCD it is _sx1). While we are using the _x1d FITS file as an example of investigating STIS data, the following methods for reading in data and viewing a spectra or other fields can be applied to the other STIS data, either calibrated or uncalibrated. For more information on STIS file naming conventions, see Types of STIS Files in the STIS Data Handbook.
Open the x1d fits file and explore its info and header:
# get information about the fits file
x1d_file = Path('./data/mastDownload/HST/odj102010/odj102010_x1d.fits')
fits.info(x1d_file)
Filename: data/mastDownload/HST/odj102010/odj102010_x1d.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 275 ()
1 SCI 1 BinTableHDU 157 1R x 19C [1I, 1I, 1024D, 1024E, 1024E, 1024E, 1024E, 1024E, 1024E, 1024I, 1E, 1E, 1I, 1E, 1E, 1E, 1E, 1024E, 1E]
The primary header that stores keyword information describing the global properties of all of the exposures in the file
# get header of the fits file
x1d_header_0 = fits.getheader(x1d_file, ext=0)
for key in ["INSTRUME", "DETECTOR", "OBSMODE", "OPT_ELEM", "CENWAVE", "PROPAPER", "TARGNAME"]:
print(f"{key}:\t{x1d_header_0[key]}")
INSTRUME: STIS
DETECTOR: FUV-MAMA
OBSMODE: ACCUM
OPT_ELEM: G140M
CENWAVE: 1540
PROPAPER: 52X0.2
TARGNAME: -ALF-CEP
You can change the keys to check for other fields and metadata, or directly print the x1d_header_0 to get all header information.
Some other metadata, such as exposure data and time, are stored in the first extension.
x1d_header_1 = fits.getheader(x1d_file, ext=1)
date = x1d_header_1["DATE-OBS"]
time = x1d_header_1["Time-OBS"]
exptime = x1d_header_1["EXPTIME"]
print(f"The data were taken on {date}, starting at {time}, with the exposure time of {exptime} seconds")
The data were taken on 2018-01-14, starting at 08:31:01, with the exposure time of 900.0 seconds
2.2 Reading table data#
The main science data is stored in the first extension of the x1d FITS file. We first read in the data as an astropy Table.
# Get data
x1d_data = Table.read(x1d_file, hdu=1)
# Display a representation of the data:
x1d_data
WARNING: UnitsWarning: 'Angstroms' did not parse as fits unit: At col 0, Unit 'Angstroms' not supported by the FITS standard. Did you mean Angstrom or angstrom? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
| SPORDER | NELEM | WAVELENGTH | GROSS | BACKGROUND | NET | FLUX | ERROR | NET_ERROR | DQ | A2CENTER | EXTRSIZE | MAXSRCH | BK1SIZE | BK2SIZE | BK1OFFST | BK2OFFST | EXTRLOCY | OFFSET |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Angstroms | Counts/s | Counts/s | Counts/s | erg / (Angstrom s cm2) | erg / (Angstrom s cm2) | Counts/s | pix | pix | pix | pix | pix | pix | pix | pix | pix | |||
| int16 | int16 | float64[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | int16[1024] | float32 | float32 | int16 | float32 | float32 | float32 | float32 | float32[1024] | float32 |
| 1 | 1024 | 1513.611692349868 .. 1567.668670092399 | 1.423279 .. 0.04860059 | 0.0008550065 .. -6.92308e-05 | 1.422424 .. 0.04866982 | 1.969044e-12 .. 9.896183e-14 | 5.50753e-14 .. 1.495939e-14 | 0.03978601 .. 0.007357089 | 2564 .. 2564 | 389.5777 | 11 | 1024 | 5 | 5 | -300 | 300 | 382.2857 .. 397.2364 | 351.8867 |
# We can also get the columns of this table:
columns = x1d_data.colnames
columns
['SPORDER',
'NELEM',
'WAVELENGTH',
'GROSS',
'BACKGROUND',
'NET',
'FLUX',
'ERROR',
'NET_ERROR',
'DQ',
'A2CENTER',
'EXTRSIZE',
'MAXSRCH',
'BK1SIZE',
'BK2SIZE',
'BK1OFFST',
'BK2OFFST',
'EXTRLOCY',
'OFFSET']
Another common way of reading in FITS data from an HDU list as “FITS_rec”:
with fits.open(x1d_file) as hdulist:
fuv_x1d_data = hdulist[1].data
type(fuv_x1d_data)
astropy.io.fits.fitsrec.FITS_rec
3. Plotting the spectrum#
3.1 Making a simple plot of the spectrum#
The actual data of each column are stored in arrays within each row with equal lengths. We collect the spectrum data and plot them with corresponding error bars.
# From the astropy table, we first get all the data we need: wavelength, flux, and error
# notice that for astropy table, the column name is case sensitive
# First-order data have data in only the 0th row, so we extract this sparse dimension.
wl, flux, err = x1d_data[0]['WAVELENGTH', 'FLUX', 'ERROR']
# Make a plot of the data, use this cell to specify the format of the plot.
matplotlib.rcParams['figure.figsize'] = [20, 7]
plt.style.use('bmh')
plt.plot(wl, flux, # the x-data & y-data
marker='.', markersize=2, markerfacecolor='w', markeredgewidth=0) # specifies the data points style
plt.fill_between(wl, flux - err, flux + err, alpha=0.5) # shade regions by width of error array
plt.title('STIS FUV Spectrum')
plt.xlabel('Wavelength (Å)')
plt.ylabel('Flux (ergs/s/cm$^2$/Å)')
plt.show()
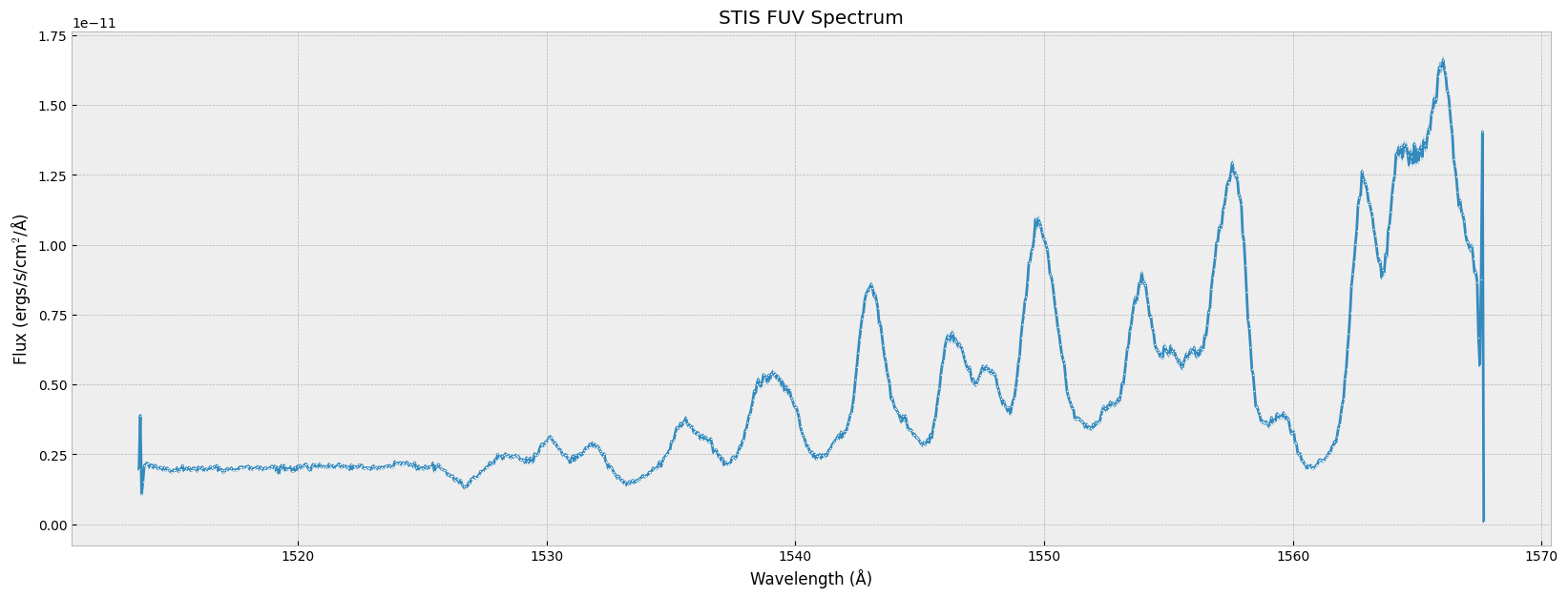
You can zoom in to a specific wavelength range
# plot the spectrum between 1540 and 1560 Å:
plt.plot(wl, flux, # the x-data & y-data
marker='.', markersize=2, markerfacecolor='w', markeredgewidth=0, # specifies the data point style
color='blue') # specifies the format of lines
plt.fill_between(wl, flux - err, flux + err, alpha=0.5) # shade regions by width of error array
plt.title('STIS FUV Spectrum')
plt.xlabel('Wavelength (Å)')
plt.ylabel("Flux (ergs/s/cm$^2$/Å)")
plt.xlim(1540, 1560)
plt.show()
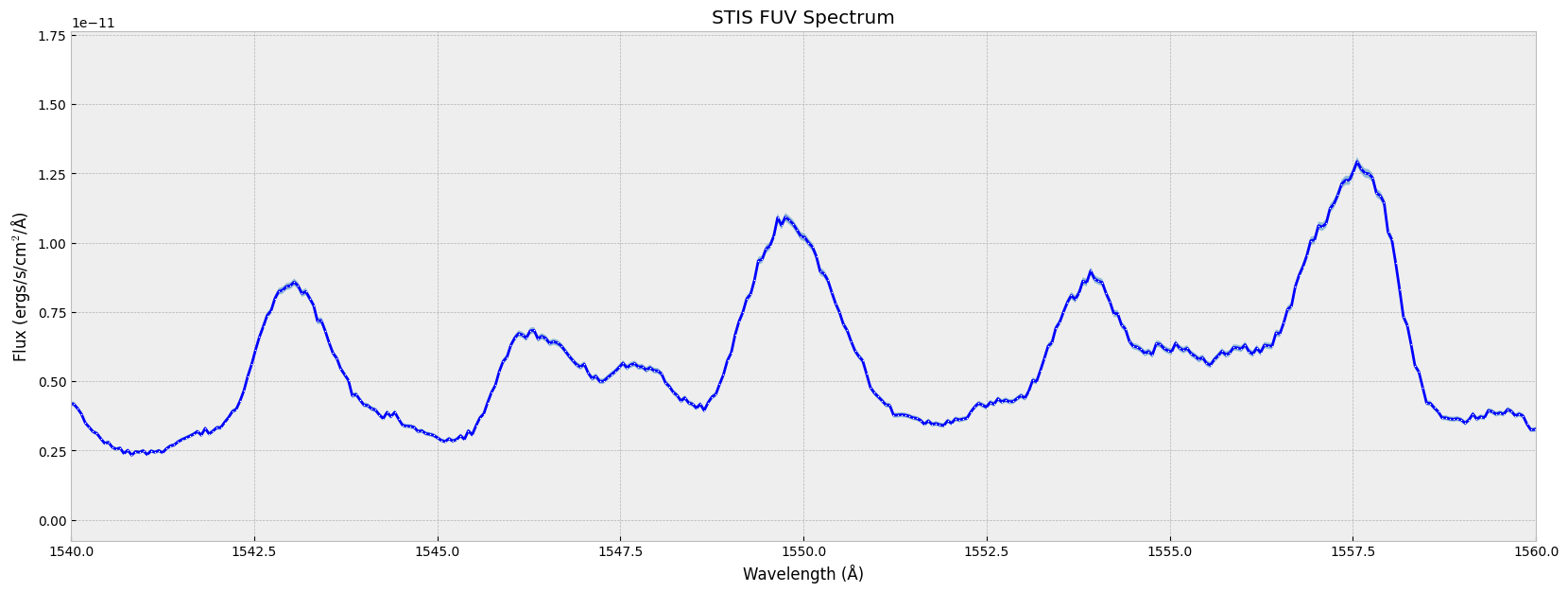
You can also plot the error bar together with the spectrum. For more errorbar styling options, see matplotlib.pyplot.errorbar
plt.errorbar(wl, flux, err, # the x-data, y-data, and y-axis error
marker='.', markersize=2, markerfacecolor='w', markeredgewidth=0, color='blue', # specifies the data points style
ecolor='dodgerblue', capsize=0.1) # specifies the format of lines and error bar
plt.title('STIS FUV Spectrum')
plt.xlabel('Wavelength (Å)')
plt.ylabel('Flux (ergs/s/cm$^2$/Å)')
plt.xlim(1540, 1560)
plt.show()
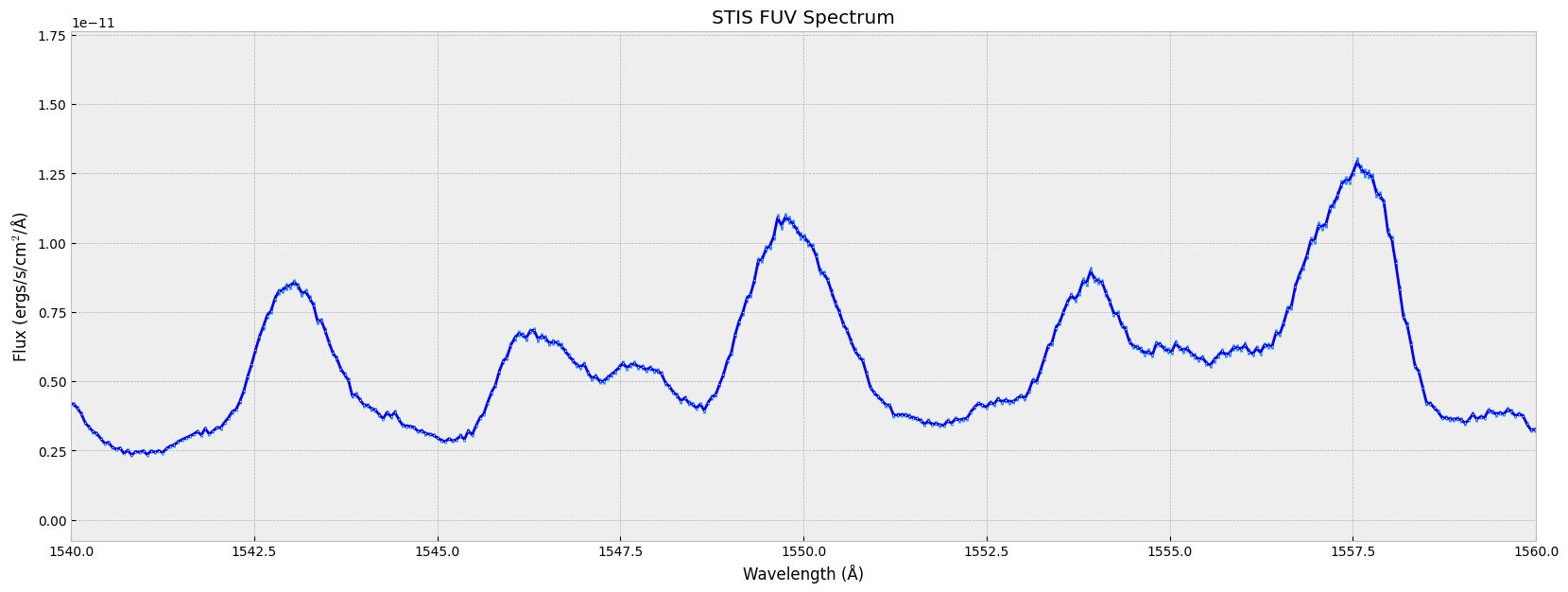
For more information on formatting the plots using matplotlib, see matplotlib.pyplot.plot, matplotlib.pyplot.errorbar.
4.Working with data quality flags#
Data quality flags are assigned to each pixel in the data quality extension. Each flag has a true (set) or false (unset) state. Flagged conditions are set as specific bits in a 16-bit integer word. For a single pixel, this allows for up to 15 data quality conditions to be flagged simultaneously, using the bitwise logical OR operation. Note that the data quality flags cannot be interpreted simply as integers but may be converted to base-2 and interpreted as flags. These flags are set and used during the course of calibration, and may likewise be interpreted and used by downstream analysis applications.
4.1 Data quality frequencies histogram#
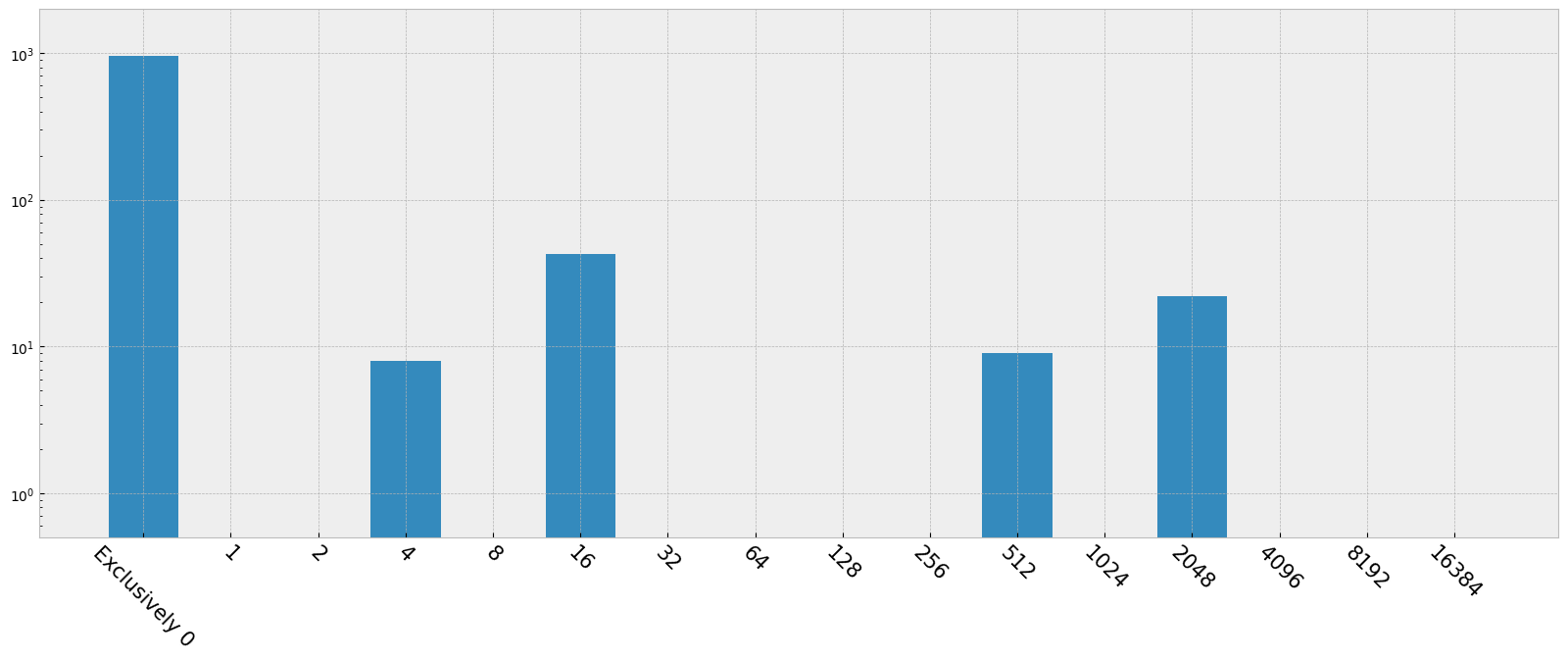
Make a histogram according to the data quality flags, and label the bins by what each data quality values actually means. More info: https://hst-docs.stsci.edu/stisdhb/chapter-2-stis-data-structure/2-5-error-and-data-quality-array
# First get the possible data quality flag values to bitwise-and with the data quality flag:
dq_flags = ['Exclusively 0'] + list(1 << np.arange(0, 15))
dq_bits = {}
# In the Table representation, the data quality flag is a masked array that "hides" the pixels
# with no data quality issue. We fill those "good points" with 0 here when counting:
dq_bits[0] = np.count_nonzero(x1d_data[0]['DQ'].filled(0) == 0) # exclusively 0
# Loop over non-zero flag values and count each:
for dq_flag in dq_flags[1:]:
dq_bits[dq_flag] = np.count_nonzero((x1d_data[0]['DQ'] & dq_flag))
dq_bits
{0: np.int64(960),
np.int64(1): np.int64(0),
np.int64(2): np.int64(0),
np.int64(4): np.int64(8),
np.int64(8): np.int64(0),
np.int64(16): np.int64(43),
np.int64(32): np.int64(0),
np.int64(64): np.int64(0),
np.int64(128): np.int64(0),
np.int64(256): np.int64(0),
np.int64(512): np.int64(9),
np.int64(1024): np.int64(0),
np.int64(2048): np.int64(22),
np.int64(4096): np.int64(0),
np.int64(8192): np.int64(0),
np.int64(16384): np.int64(0)}
# Assign the meaning of each data quality and make a histogram
meanings = {
0: "No Anomalies",
1: "Error in the Reed Solomon decoding",
2: "Lost data replaced by fill values",
3: "Bad detector pixel (e.g., bad column or row, mixed science and bias for overscan, or beyond aperture)",
4: "Data masked by occulting bar",
5: "Pixel having dark rate > 5 σ times the median dark level",
6: "Large blemish, depth > 40% of the normalized p-flat (repeller wire)",
7: "Vignetted pixel",
8: "Pixel in the overscan region",
9: "Saturated pixel, count rate at 90% of max possible—local non-linearity turns over and is multi-valued; "
"pixels within 10% of turnover and all pixels within 4 pixels of that pixel are flagged.",
10: "Bad pixel in reference file",
11: "Small blemish, depth between 40% and 70% of the normalized flat. Applies to MAMA and CCD p-flats",
12: ">30% of background pixels rejected by sigma-clip, or flagged, during 1-D spectral extraction",
13: "Extracted flux affected by bad input data",
14: "Data rejected in input pixel during image combination for cosmic ray rejection",
15: "Extracted flux not CTI corrected because gross counts are ≤ 0"}
plt.bar(meanings.keys(), height=dq_bits.values(), tick_label=dq_flags)
plt.gca().set_xticklabels(dq_flags, rotation=-45, fontsize=15)
plt.yscale('log')
plt.ylim(0.5, 2000)
plt.show()
bits = [f'{0:015b}'] + [f'{1 << x:015b}' for x in range(0, 15)]
dq_table = pd.DataFrame({
"Flag Value": dq_flags,
"Bit Setting": bits,
"Quality Condition Indicated": meanings.values()})
dq_table.set_index('Flag Value', drop=True, inplace=True)
pd.set_option('display.max_colwidth', None)
display(dq_table)
| Bit Setting | Quality Condition Indicated | |
|---|---|---|
| Flag Value | ||
| Exclusively 0 | 000000000000000 | No Anomalies |
| 1 | 000000000000001 | Error in the Reed Solomon decoding |
| 2 | 000000000000010 | Lost data replaced by fill values |
| 4 | 000000000000100 | Bad detector pixel (e.g., bad column or row, mixed science and bias for overscan, or beyond aperture) |
| 8 | 000000000001000 | Data masked by occulting bar |
| 16 | 000000000010000 | Pixel having dark rate > 5 σ times the median dark level |
| 32 | 000000000100000 | Large blemish, depth > 40% of the normalized p-flat (repeller wire) |
| 64 | 000000001000000 | Vignetted pixel |
| 128 | 000000010000000 | Pixel in the overscan region |
| 256 | 000000100000000 | Saturated pixel, count rate at 90% of max possible—local non-linearity turns over and is multi-valued; pixels within 10% of turnover and all pixels within 4 pixels of that pixel are flagged. |
| 512 | 000001000000000 | Bad pixel in reference file |
| 1024 | 000010000000000 | Small blemish, depth between 40% and 70% of the normalized flat. Applies to MAMA and CCD p-flats |
| 2048 | 000100000000000 | >30% of background pixels rejected by sigma-clip, or flagged, during 1-D spectral extraction |
| 4096 | 001000000000000 | Extracted flux affected by bad input data |
| 8192 | 010000000000000 | Data rejected in input pixel during image combination for cosmic ray rejection |
| 16384 | 100000000000000 | Extracted flux not CTI corrected because gross counts are ≤ 0 |
4.2 Removing “Serious Data Quality Flags”#
Through the calibaration pipeline, some data qualities are marked “serious”. The value of serious data qualities are marked through “SDQFLAGS”. We can decompose that value according to the bits in order to see the specific data quality flags that are marked serious.
sdqFlags_fuv = fits.getval(x1d_file, ext=1, keyword="SDQFLAGS")
print(f'The SDQFLAGS is {sdqFlags_fuv}, which is in binary {sdqFlags_fuv:015b},')
print('Therefore the following data qualities are marked "serious":')
for i in range(15):
if 2**i & sdqFlags_fuv:
print(f'\t{i+1:2.0f}: ' + meanings[i+1])
The SDQFLAGS is 31743, which is in binary 111101111111111,
Therefore the following data qualities are marked "serious":
1: Error in the Reed Solomon decoding
2: Lost data replaced by fill values
3: Bad detector pixel (e.g., bad column or row, mixed science and bias for overscan, or beyond aperture)
4: Data masked by occulting bar
5: Pixel having dark rate > 5 σ times the median dark level
6: Large blemish, depth > 40% of the normalized p-flat (repeller wire)
7: Vignetted pixel
8: Pixel in the overscan region
9: Saturated pixel, count rate at 90% of max possible—local non-linearity turns over and is multi-valued; pixels within 10% of turnover and all pixels within 4 pixels of that pixel are flagged.
10: Bad pixel in reference file
12: >30% of background pixels rejected by sigma-clip, or flagged, during 1-D spectral extraction
13: Extracted flux affected by bad input data
14: Data rejected in input pixel during image combination for cosmic ray rejection
15: Extracted flux not CTI corrected because gross counts are ≤ 0
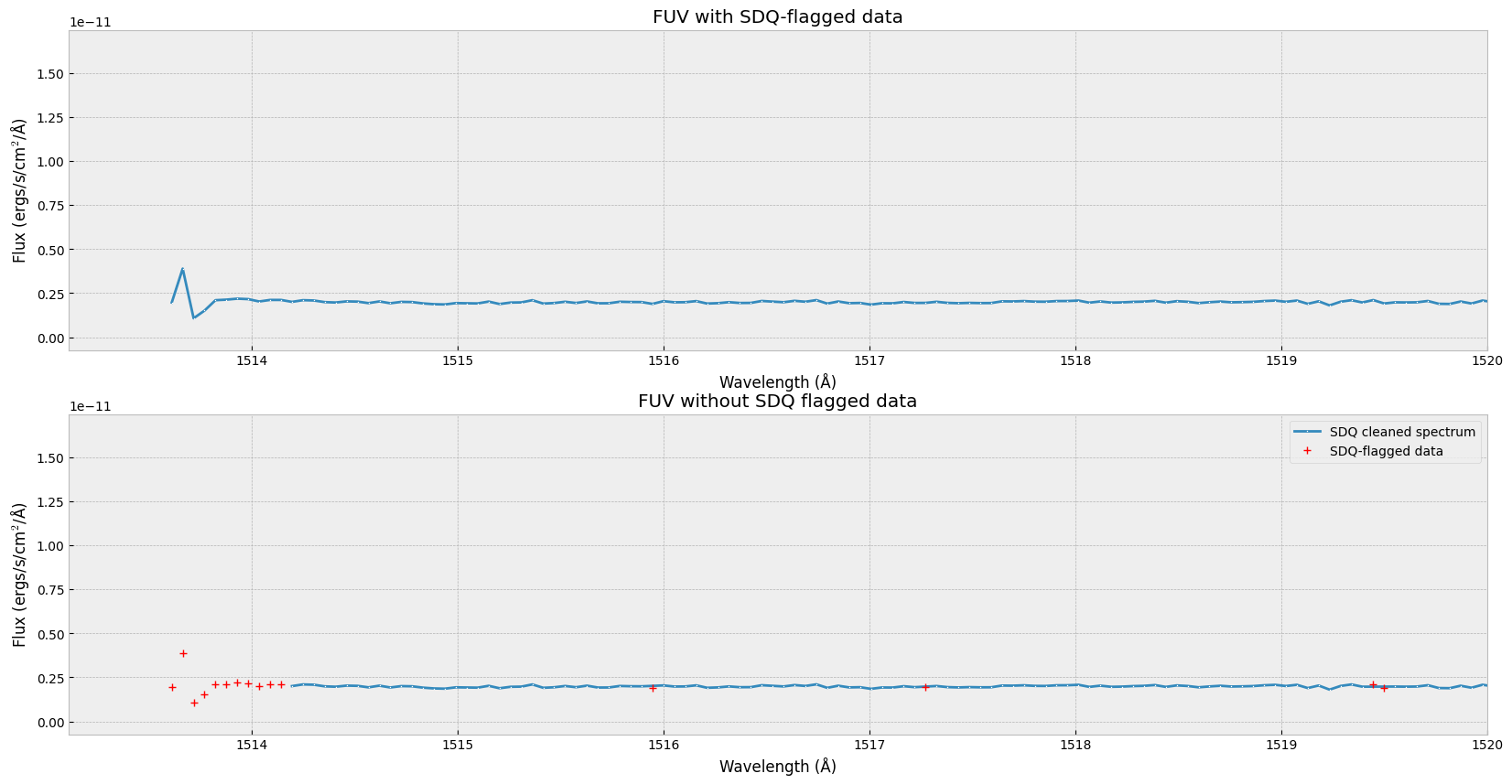
We can now remove the data points with SDQ flags. In the plots below, the first one is the same as the spectrum in which we do not handle the SDQ flags. The second one is the spectrum with SDQ flagged data removed, and the SDQ flagged data points are marked with red ‘+’.
fig, axes = plt.subplots(2, 1)
fig.set_size_inches(20, 10)
plt.style.use('bmh')
# First zoom in to the region where SDQ got removed
wl, flux, err, dq = x1d_data[0]['WAVELENGTH', 'FLUX', 'ERROR', 'DQ']
# Filter the datapoints to where there are no serious DQ flags
mask_noSDQ = (dq & sdqFlags_fuv) == 0
# get the spectrum without SDQ using the mask we just created
wvln_noSDQ, flux_noSDQ, err_noSDQ = wl[mask_noSDQ], flux[mask_noSDQ], err[mask_noSDQ]
# inverse the _noSDQ mask to collect the data points with SDQ flags
wvln_SDQ, flux_SDQ = wl[~mask_noSDQ], flux[~mask_noSDQ] # apply the bitwise-not of mask
# plot1: the spectrum with SDQ flagged data included
# plt.subplot(2,1,1)
axes[0].plot(wl, flux, # the x-data, y-data
marker='.', markersize=2, markerfacecolor='w', markeredgewidth=0) # specifies the data points style
# plot2: plot the spectrum without SDQ flagged data, then mark the SDQ data points with +
# Plot the filtered datapoints
axes[1].plot(wvln_noSDQ, flux_noSDQ, # the x-data, y-data
marker='.', markersize=2, markerfacecolor='w', markeredgewidth=0, label="SDQ cleaned spectrum")
axes[1].plot(wvln_SDQ, flux_SDQ, 'r+', label='SDQ-flagged data')
axes[1].legend(loc='best')
# Format the figures:
axes[0].set_title('FUV with SDQ-flagged data')
axes[1].set_title('FUV without SDQ flagged data')
for ax in axes:
ax.set_xlabel('Wavelength (Å)')
ax.set_ylabel('Flux (ergs/s/cm$^2$/Å)')
ax.set_xlim(wl.min() - 0.5, 1520) # zoom in on left edge
5. Visualizing STIS Image#
The STIS images are stored as two-dimensional arrays in FITS image extension files. For more information on STIS image files and extension, see STIS FITS Image Extension Files
5.1 Exploring image file structure#
The rectified, wavelength and flux calibrated first order spectra or Geometrically corrected imaging data is stored in the fits file with the x2d extension (note that for the CCD the similar extension is sx2). Similar to what we did to the x1d file, we first open the fits file to explore its file structure.
# read in the x2d file and get its info
x2d_file = Path('./data/mastDownload/HST/odj102010/odj102010_x2d.fits')
fits.info(x2d_file)
Filename: data/mastDownload/HST/odj102010/odj102010_x2d.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 278 ()
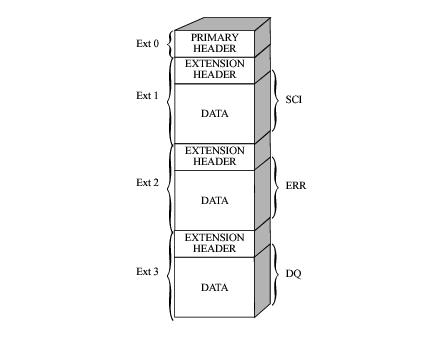
1 SCI 1 ImageHDU 121 (1201, 1201) float32
2 ERR 1 ImageHDU 61 (1201, 1201) float32
3 DQ 1 ImageHDU 44 (1201, 1201) int16
The first, of extension type SCI, stores the science count/flux values.
The second, of extension type ERR, contains the statistical errors, which are propagated through the calibration process. It is unpopulated in raw data files.
The third, of extension type DQ, stores the data quality values, which flag suspect pixels in the corresponding SCI data. It is unpopulated in raw data files.
Similarly, we can also get the header from this FITS file to see the scientific metadata.
# get header of the fits file
x2d_header = fits.getheader(x2d_file, ext=0)
x2d_header[:15] # additional lines are not displayed here
SIMPLE = T / file does conform to FITS standard
BITPIX = 16 / number of bits per data pixel
NAXIS = 0 / number of data axes
EXTEND = T / FITS dataset may contain extensions
COMMENT FITS (Flexible Image Transport System) format is defined in 'Astronomy
COMMENT and Astrophysics', volume 376, page 359; bibcode: 2001A&A...376..359H
ORIGIN = 'HSTIO/CFITSIO March 2010' / FITS file originator
DATE = '2025-09-30' / date this file was written (yyyy-mm-dd)
NEXTEND = 3 / Number of standard extensions
FILENAME= 'odj102010_x2d.fits' / name of file
FILETYPE= 'SCI ' / type of data found in data file
TELESCOP= 'HST' / telescope used to acquire data
BSIDEOPS= 'FALSE' / Post-2021-07-15 B-Side Operations Indicator
INSTRUME= 'STIS ' / identifier for instrument used to acquire data
5.2 Showing the image#
Now we collect the science image data from the fits file and show the image.
# get data as a numpy array
with fits.open(x2d_file) as hdu_list:
x2d_data = hdu_list[1].data
Make a histogram of the magnitude of the image data so that we have a general idea on the distribution. Knowing the distribution is essential for us to normalize the data when showing the image.
# remove infinities and NaNs from the data when making the histogram
filtered_data = [v for row in np.log10(x2d_data) for v in row if not (np.isinf(v) or np.isnan(v))]
fig, ax = plt.subplots(1, 1)
fig.set_size_inches(15, 5)
ax.hist(filtered_data, range=[-20.5, -8.5], bins=12)
ax.set_xlabel('Magnitude order of _x2d image data')
ax.set_ylabel('count')
ax.set_title('_x2d Data Magnitude Order frequencies')
/tmp/ipykernel_2949/3318306081.py:2: RuntimeWarning: divide by zero encountered in log10
filtered_data = [v for row in np.log10(x2d_data) for v in row if not (np.isinf(v) or np.isnan(v))]
/tmp/ipykernel_2949/3318306081.py:2: RuntimeWarning: invalid value encountered in log10
filtered_data = [v for row in np.log10(x2d_data) for v in row if not (np.isinf(v) or np.isnan(v))]
Text(0.5, 1.0, '_x2d Data Magnitude Order frequencies')
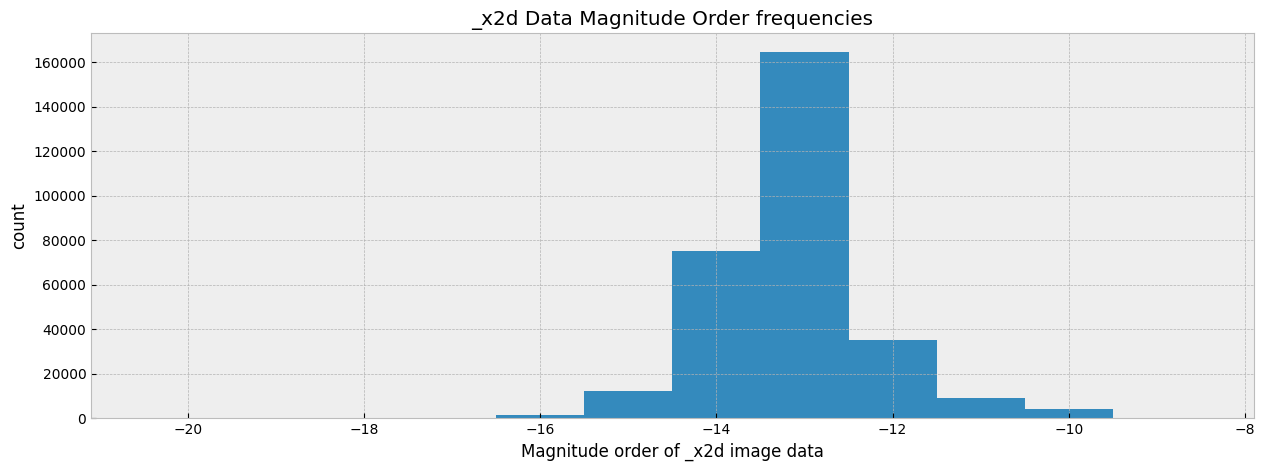
When showing the image, we normalize the color of each pixel to a specific range through vmin and vmax to make the features of image clear. These values typically matches the magnitude of the x2d data according to the histogram above, but can be experimented and changed to bring out features at different levels.
# show the image
matplotlib.rcParams['figure.figsize'] = (10, 10)
plt.imshow(x2d_data, origin='lower', vmin=0, vmax=1e-13, cmap="viridis")
<matplotlib.image.AxesImage at 0x7ff358194c10>
For more color map and normalizing options, see: Choosing Colormaps in Matplotlib, matplotlib.pyplot.imshow.
5.3 Removing serious data quality pixels#
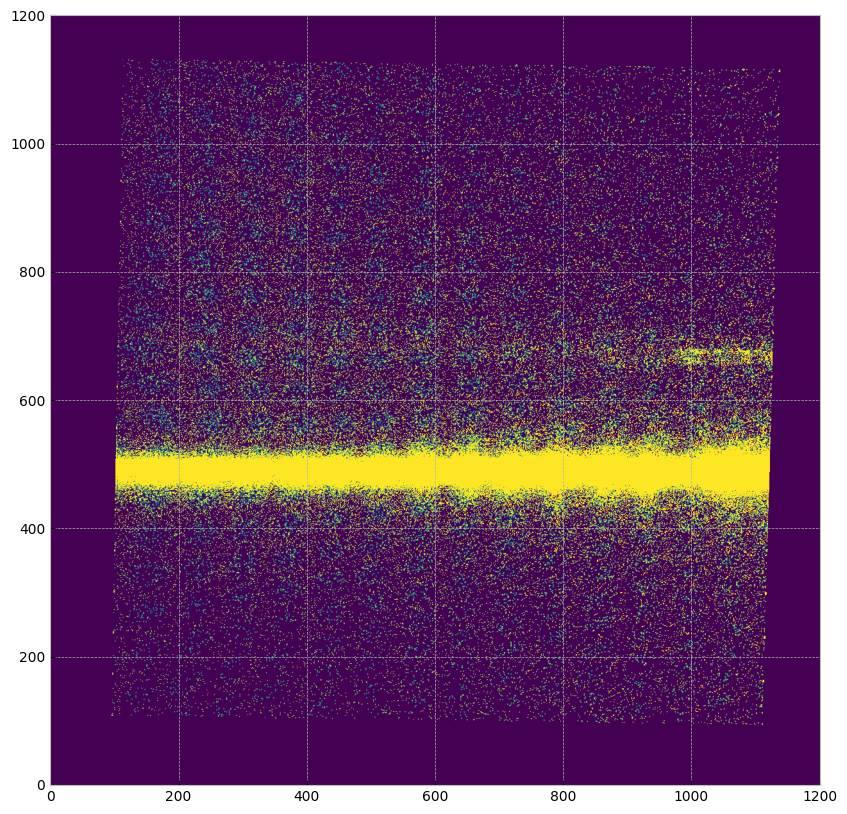
Now we are going to remove the pixels with series data quality flags as described above. The removed pixels will appear grey like the background. The horizontal stripes of removed data are from the bar (top) and repeller wire shadow (middle).
# get the serious data quality flag
sdqFlags_fuv = fits.getheader(x2d_file, 1)["SDQFLAGS"]
# get data quality flags of each pixels
with fits.open(x2d_file) as hdu_list:
x2d_dq = hdu_list[3].data
# create a mask of bad pixels and set them to nan
def check_dq(dq):
return bool(dq & sdqFlags_fuv)
mask = np.vectorize(check_dq)(x2d_dq)
x2d_mask = np.ma.array(x2d_data, mask=mask, fill_value=np.nan)
# plot the image
plt.imshow(x2d_mask, origin='lower', vmin=0, vmax=1e-13, cmap="viridis")
<matplotlib.image.AxesImage at 0x7ff34e4f72d0>
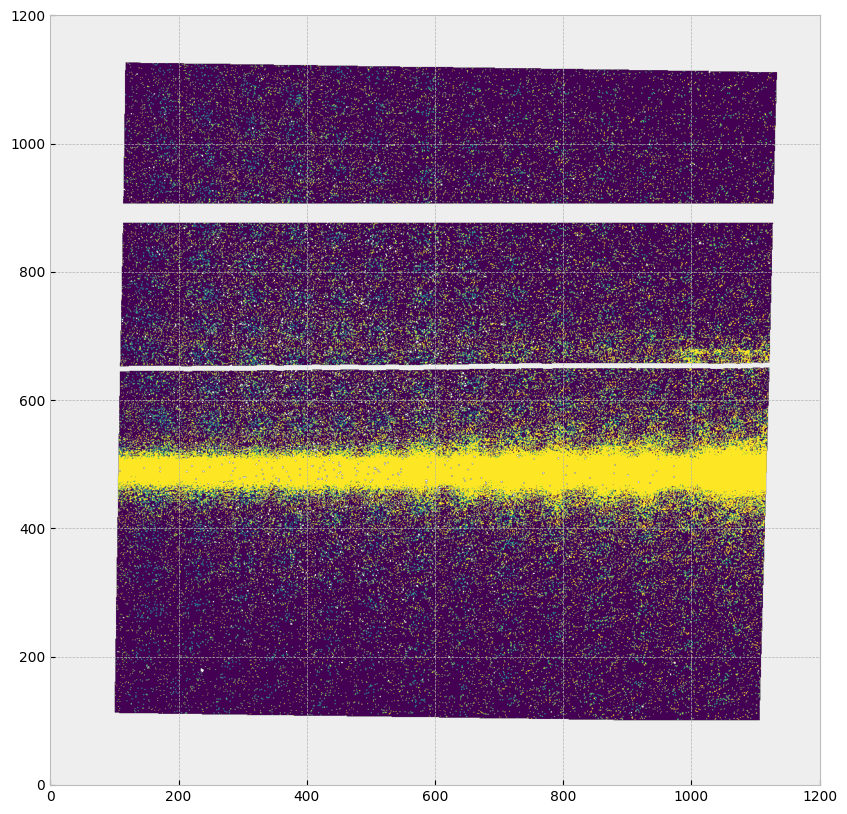
6.Working with Time-Tag data#
The MAMA detecters have a unique Time-Tag mode besides ACCUM mode. TIME-TAG mode is used for high-time-resolution spectroscopy and imaging in the UV. In TIME-TAG mode, the position and detection time of every photon is recorded in an event list. The Time-Tag mode operation for the MAMA detectors can be found at: MAMA TIME-TAG Mode.
In TIME-TAG mode, the position and detection time of every photon is recorded in an event list. First download the _tag data:
%%capture --no-stderr --no-display
# Search target objscy by obs_id
target = Observations.query_criteria(obs_id='odgxt9010')
# get a list of files assiciated with that target
NUV_list = Observations.get_product_list(target)
# Download only the SCIENCE fits files
Observations.download_products(NUV_list, extension='fits', download_dir=str(datadir))
| Local Path | Status | Message | URL |
|---|---|---|---|
| str119 | str8 | object | object |
| data/mastDownload/HST/odgxt9010/odgxt9010_jif.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_jit.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_jwf.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_jwt.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_spt.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_tag.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_trl.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_wav.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_wsp.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_asn.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_raw.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_x2d.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_flt.fits | COMPLETE | None | None |
| data/mastDownload/HST/odgxt9010/odgxt9010_x1d.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_stis_v-t-cra_odgx/hst_15070_stis_v-t-cra_g230m_odgx_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_stis_v-t-cra_odgx/hst_15070_stis_v-t-cra_g230m_odgx09_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_stis_v-t-cra_odgx/hst_15070_stis_v-t-cra_g230m_odgxc9_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_stis_v-t-cra_odgx/hst_15070_stis_v-t-cra_g230m_odgxt9_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_cos-stis_v-t-cra_ldgx/hst_15070_cos-stis_v-t-cra_g130m-g160m-g230m_ldgx_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_15070_cos-stis_v-t-cra_ldgx/hst_15070_cos-stis_v-t-cra_g130m-g230m_ldgx_cspec.fits | COMPLETE | None | None |
6.1 Investigating the _tag data#
# get info about the tag extension fits file
tag = Path("./data/mastDownload/HST/odgxt9010/odgxt9010_tag.fits")
fits.info(tag)
Filename: data/mastDownload/HST/odgxt9010/odgxt9010_tag.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 219 ()
1 EVENTS 1 BinTableHDU 148 4802131R x 4C [1J, 1I, 1I, 1I]
2 GTI 1 BinTableHDU 22 1R x 2C [1D, 1D]
The _tag fits file has two binary table extensions: EVENTS and GTI (Good Time Interval).
# get header of the EVENTS extension
# print only the TIMETAG EVENTS TABLE COLUMNS (line 130-147)
fits.getheader(tag, 1)[130:147]
SHIFTA2 = 0.000000 / Spectrum shift in AXIS2 calculated from WAVECAL
/ TIMETAG EVENTS TABLE COLUMNS
TTYPE1 = 'TIME ' / event clock time
TFORM1 = '1J ' / data format for TIME: 32-bit integer
TUNIT1 = 'seconds ' / units for TIME: seconds
TSCAL1 = 0.000125 / scale factor to convert s/c clock ticks to sec
TZERO1 = 0.0 / TIME zero point: starting time of obs.
TTYPE2 = 'AXIS1 ' / Doppler corrected axis 1 detector coordinate
TFORM2 = '1I ' / data format for AXIS1: 16-bit integer
TUNIT2 = 'pixels ' / physical units for AXIS1: pixels
TTYPE3 = 'AXIS2 ' / axis 2 detector coordinate
TFORM3 = '1I ' / data format for AXIS2: 16-bit integer
TUNIT3 = 'pixels ' / physical units for AXIS2: pixels
TTYPE4 = 'DETAXIS1' / raw 1 detector coordinate
TFORM4 = '1I ' / data format for DETAXIS1: 16-bit integer
Columns in the EVENTS extension:
TIME: the time each event was recorded relative to the start time
AXIS1: pixel coordinate along the spectral axis with the corretion term on Doppler shifts
AXIS2: pixel coordinate along the spatial axis
DETAXIS1: pixel coordinate along the spectral axis without the corretion term on Doppler shifts
# get header of the GTI extension
fits.getheader(tag, 2)
XTENSION= 'BINTABLE' / extension type
BITPIX = 8 / bits per data value
NAXIS = 2 / number of data axes
NAXIS1 = 16 / length of first data axis
NAXIS2 = 1 / length of second data axis
PCOUNT = 0 / number of group parameters
GCOUNT = 1 / number of groups
TFIELDS = 2 / number of fields in each table row
INHERIT = T / inherit the primary header
EXTNAME = 'GTI ' / extension name
EXTVER = 1 / extension version number
ROOTNAME= 'odgxt9010 ' / rootname of the observation set
EXPNAME = 'odgxt9rfq ' / exposure identifier
/ TIMETAG GTI TABLE COLUMN DESCRIPTORS
TTYPE1 = 'START ' / start of good time interval
TFORM1 = '1D ' / data format for START: REAL*8
TUNIT1 = 'seconds ' / physical units for START
TTYPE2 = 'STOP ' / end of good time interval
TFORM2 = '1D ' / data format for STOP: REAL*8
TUNIT2 = 'seconds ' / physical units for STOP
Table.read(tag, 2)
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
| START | STOP |
|---|---|
| seconds | seconds |
| float64 | float64 |
| 0.022625 | 2380.222 |
Columns in the GTI extension:
START: start of good time interval
STOP: end of good time interval
Now we make a time series plot of the total flux over all wavelengths to see how the flux changes over the time interval:
# read the events data as a pandas dataframe for easier manipulation
events = Table.read(tag, 1).to_pandas()
# get the good time interval from the GTI extension
start, stop = Table.read(tag, 2)["START"][0], Table.read(tag, 2)["STOP"][0]
# group the events by time with bin = 3 seconds
time = np.arange(int(start), int(stop), 3)
ind = np.digitize(events['TIME'], time)
total_flux = events.groupby(ind).count()["TIME"]
# plot the flux as a function of time
# notice here that the unit of flux is counts since we are counting the number of events in a time series
matplotlib.rcParams['figure.figsize'] = (20, 7)
plt.plot(time, total_flux, marker=".", mfc="w")
plt.xlabel("Time [s]")
plt.ylabel("Total Flux [counts]")
plt.show()
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'pixels' did not parse as fits unit: At col 0, Unit 'pixels' not supported by the FITS standard. Did you mean pixel? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'pixels' did not parse as fits unit: At col 0, Unit 'pixels' not supported by the FITS standard. Did you mean pixel? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'pixels' did not parse as fits unit: At col 0, Unit 'pixels' not supported by the FITS standard. Did you mean pixel? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'seconds' did not parse as fits unit: At col 0, Unit 'seconds' not supported by the FITS standard. If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
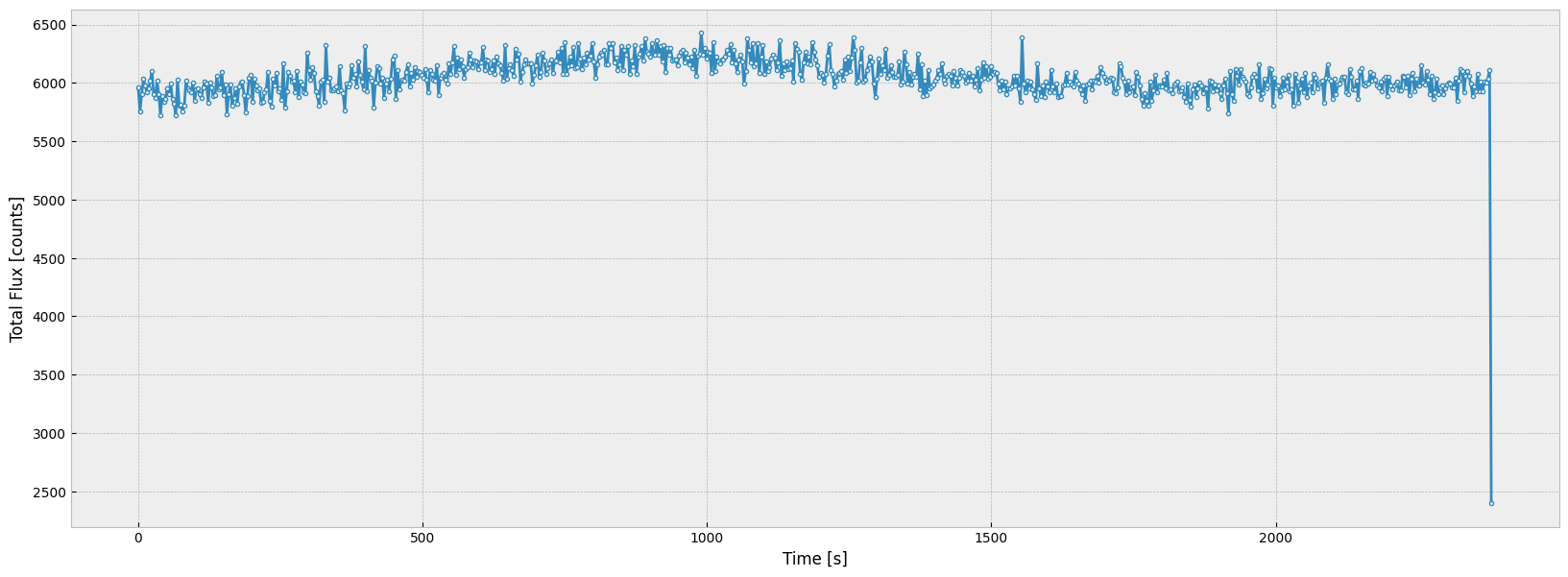
As the plot shows, though the total flux fluctuates, it is roughly a constant over the good time interval.
6.2 Converting Time_Tag into ACCUM image#
Time tag data can be converted into ACCUM image using the inttag method in stistools. More information: inttag
# define the output file
accum = "./data/mastDownload/HST/odgxt9010/odgxt9010_accum.fits"
# convert Time_Tag into ACCUM
# the first parameter is the path to the _tag fits file, the second parameter is the output directory
stistools.inttag.inttag(tag, accum)
imset: 1, start: 0.022625, stop: 2380.222, exposure time: 2380.199375
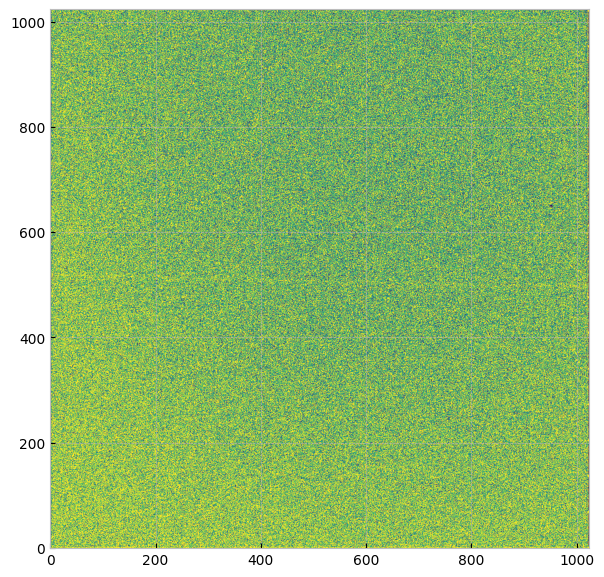
Then the output file is in the same structure as a STIS image fits file, which we can open and plot in the same way we explored above:
with fits.open(accum) as hdul:
im = hdul[1].data
plt.imshow(im, origin='lower', vmin=0, vmax=6, cmap="viridis")
<matplotlib.image.AxesImage at 0x7ff35b322510>
inttag can be run to produce multiple output imsets: rcount specifies the number of imsets, imcrements specifies the time interval for each imsets in seconds
stistools.inttag.inttag(tag, accum, rcount=3, increment=700)
fits.info(accum)
imset: 1, start: 0.022625, stop: 700.022625, exposure time: 700.0
imset: 2, start: 700.022625, stop: 1400.022625, exposure time: 700.0000000000001
imset: 3, start: 1400.022625, stop: 2100.022625, exposure time: 700.0
Filename: ./data/mastDownload/HST/odgxt9010/odgxt9010_accum.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 219 ()
1 SCI 1 ImageHDU 139 (1024, 1024) float64
2 ERR 1 ImageHDU 140 ()
3 DQ 1 ImageHDU 140 ()
4 SCI 2 ImageHDU 139 (1024, 1024) float64
5 ERR 2 ImageHDU 140 ()
6 DQ 2 ImageHDU 140 ()
7 SCI 3 ImageHDU 139 (1024, 1024) float64
8 ERR 3 ImageHDU 140 ()
9 DQ 3 ImageHDU 140 ()
Compare the 3 accum images produced by inttag, using the same scale and colormap:
for i in range(1, 8, 3):
with fits.open(accum) as hdul:
im = hdul[i].data
plt.subplot(1, 3, int(i/3)+1)
plt.imshow(im, origin='lower', vmin=0, vmax=2, cmap="viridis")
plt.title("extension {}".format(i))
plt.tight_layout()
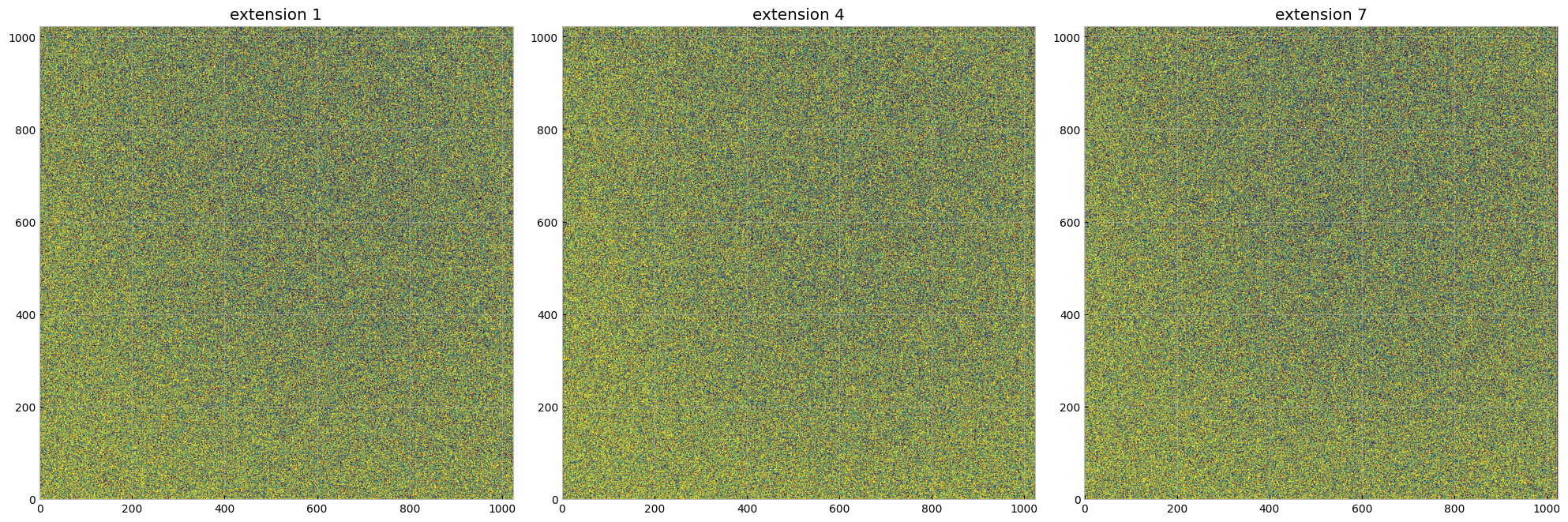
The output file is a series of extensions with each imset having a SCI, ERR, and DQ extension, as shown above.
7.Working with STIS Echelle data#
The STIS Echelle mode uses diffraction and interference to separate a spectrum into different spectral orders (the keyword in the headers is SPORDER), with each spectral order covering different wavelength range. The echelle were designed to maximize the spectral range covered in a single echellogram. For more information, see Echelle Spectroscopy in the Ultraviolet.
We first download data taken with an echelle:
%%capture --no-stderr --no-display
# Search target objscy by obs_id
target = Observations.query_criteria(obs_id='OCTX01030')
# get a list of files assiciated with that target
NUV_list = Observations.get_product_list(target)
# Download only the SCIENCE fits files
Observations.download_products(NUV_list, productType="SCIENCE", extension='fits', download_dir=str(datadir))
| Local Path | Status | Message | URL |
|---|---|---|---|
| str115 | str8 | object | object |
| data/mastDownload/HST/octx01030/octx01030_raw.fits | COMPLETE | None | None |
| data/mastDownload/HST/octx01030/octx01030_flt.fits | COMPLETE | None | None |
| data/mastDownload/HST/octx01030/octx01030_x1d.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_14094_stis_-rho-cnc_octx/hst_14094_stis_-rho-cnc_e230m_octx_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_14094_stis_-rho-cnc_octx/hst_14094_stis_-rho-cnc_e230m_octx01_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_14094_stis_-rho-cnc_octx/hst_14094_stis_-rho-cnc_e230m_octx03_cspec.fits | COMPLETE | None | None |
| data/mastDownload/HST/hst_hasp_14094_cos-stis_-rho-cnc_lctx/hst_14094_cos-stis_-rho-cnc_g130m-e230m_lctx_cspec.fits | COMPLETE | None | None |
7.1 Showing Echelle Image#
Open the _flt (Flat-fielded science) image in the same way we open other image files, and show the image:
flt_file = "./data/mastDownload/HST/octx01030/octx01030_flt.fits"
with fits.open(flt_file) as hdu:
image = hdu[1].data
plt.imshow(image, origin='lower', vmin=0, vmax=1, cmap="viridis")
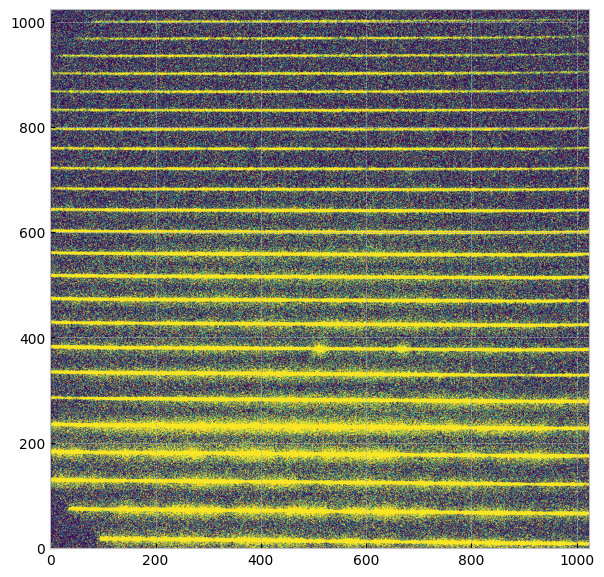
As shown in the image above, there are multiple horizontal bands with different spectral orders. Each spectral order also has distinct wavelength range, which we will explore in the next section.
7.2 Plotting Echelle Spectrum#
We first read in the _x1d data as an astropy table. Notice that when we read in the FUV _x1d data in section2.2, the table has a single row with SPORDER = 1. But for echelle mode data, the data is separated into multiple rows, with each row having a distinct order.
echelle_file = "./data/mastDownload/HST/octx01030/octx01030_x1d.fits"
echelle_data = Table.read(echelle_file, 1)
echelle_data
WARNING: UnitsWarning: 'Angstroms' did not parse as fits unit: At col 0, Unit 'Angstroms' not supported by the FITS standard. Did you mean Angstrom or angstrom? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
| SPORDER | NELEM | WAVELENGTH | GROSS | BACKGROUND | NET | FLUX | ERROR | NET_ERROR | DQ | A2CENTER | EXTRSIZE | MAXSRCH | BK1SIZE | BK2SIZE | BK1OFFST | BK2OFFST | EXTRLOCY | OFFSET |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Angstroms | Counts/s | Counts/s | Counts/s | erg / (Angstrom s cm2) | erg / (Angstrom s cm2) | Counts/s | pix | pix | pix | pix | pix | pix | pix | pix | pix | |||
| int16 | int16 | float64[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | float32[1024] | int16[1024] | float32 | float32 | int16 | float32 | float32 | float32 | float32 | float32[1024] | float32 |
| 66 | 1024 | 3067.682086787238 .. 3119.012536824325 | -0.0002752173 .. 0.4856005 | 0.006653455 .. 0.003303319 | -0.006928673 .. 0.4822972 | -2.024925e-14 .. 2.961075e-12 | 6.728192e-17 .. 4.64744e-13 | 2.302181e-05 .. 0.07569709 | 2628 .. 2564 | 14.40743 | 7 | 10 | 5 | 5 | 28.01995 | -28.77343 | 19.4528 .. 9.476286 | 0.6031421 |
| 67 | 1024 | 3021.857426990342 .. 3072.451048939282 | -0.0002702794 .. 0.832027 | 0.004700221 .. 0.000911057 | -0.0049705 .. 0.831116 | -1.016979e-14 .. 3.081053e-12 | 4.684728e-17 .. 3.703201e-13 | 2.289667e-05 .. 0.09989408 | 2628 .. 2580 | 71.19328 | 7 | 10 | 5 | 5 | 27.28738 | -28.01995 | 75.9551 .. 66.73717 | 0.6031421 |
| 68 | 1024 | 2977.380919743117 .. 3027.257338436938 | -0.008085039 .. 0.4208066 | 0.01313397 .. 0.002160907 | -0.02121901 .. 0.4186457 | -3.668029e-14 .. 1.092327e-12 | 2.15723e-16 .. 1.857869e-13 | 0.0001247926 .. 0.07120476 | 2580 .. 2564 | 126.3682 | 7 | 10 | 5 | 5 | 26.57536 | -27.28738 | 131.2862 .. 122.4106 | 0.6031421 |
| 69 | 1024 | 2934.193937177996 .. 2983.372006753561 | -0.0002734362 .. 0.9764332 | 0.01347408 .. 0.002397537 | -0.01374752 .. 0.9740357 | -2.182534e-14 .. 2.149785e-12 | 3.654991e-17 .. 2.292959e-13 | 2.302235e-05 .. 0.1038906 | 2564 .. 2564 | 179.9044 | 7 | 10 | 5 | 5 | 25.88353 | -26.57536 | 184.4561 .. 176.3899 | 0.5168139 |
| 70 | 1024 | 2892.241202224274 .. 2940.739045949361 | -0.00034477 .. 1.414328 | 0.01000156 .. 0.00384891 | -0.01034633 .. 1.410479 | -1.473246e-14 .. 3.039156e-12 | 3.682563e-17 .. 2.665639e-13 | 2.586195e-05 .. 0.1237129 | 2564 .. 2564 | 232.084 | 7 | 10 | 5 | 5 | 25.21152 | -25.88353 | 236.3518 .. 228.9839 | 0.5693105 |
| 71 | 1024 | 2851.470552596696 .. 2899.305600156981 | -0.0002438011 .. 1.354866 | 0.002504314 .. 0.005019426 | -0.002748115 .. 1.349846 | -3.943758e-15 .. 2.763269e-12 | 3.14009e-17 .. 2.479534e-13 | 2.188098e-05 .. 0.1211243 | 2564 .. 2564 | 282.7351 | 7 | 10 | 5 | 5 | 24.55895 | -25.21152 | 286.8198 .. 280.1042 | 0.5335838 |
| 72 | 1024 | 2811.832724454624 .. 2859.021746892561 | -0.0002620455 .. 0.3707643 | 0.005110799 .. 0.00183624 | -0.005372844 .. 0.3689281 | -7.33356e-15 .. 7.601616e-13 | 3.062765e-17 .. 1.381673e-13 | 2.243897e-05 .. 0.06705654 | 2564 .. 2564 | 332.0811 | 7 | 10 | 5 | 5 | 23.92547 | -24.55895 | 335.8839 .. 329.9276 | 0.5793881 |
| 73 | 1024 | 2773.281153845325 .. 2819.840296326157 | -0.007080508 .. 0.7288955 | 0.005753997 .. 0.002980709 | -0.01283451 .. 0.7259148 | -1.82008e-14 .. 1.55503e-12 | 1.656346e-16 .. 1.939602e-13 | 0.0001167992 .. 0.09054399 | 2580 .. 2564 | 380.1232 | 7 | 10 | 5 | 5 | 23.31071 | -23.92547 | 383.655 .. 378.4149 | 0.6562741 |
| 74 | 1024 | 2735.771794248858 .. 2781.716606813955 | -0.0002173291 .. 0.3042348 | 0.003891864 .. 0.001904726 | -0.004109193 .. 0.3023301 | -6.061262e-15 .. 6.815853e-13 | 3.030601e-17 .. 1.387218e-13 | 2.054576e-05 .. 0.06153269 | 2564 .. 2564 | 426.7452 | 7 | 10 | 5 | 5 | 22.7143 | -23.31071 | 430.1284 .. 425.6361 | 0.5977532 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 81 | 1024 | 2499.145134153861 .. 2541.180702643316 | -0.000323896 .. 0.1212655 | 0.001022041 .. 0.0004910827 | -0.001345937 .. 0.1207744 | -2.612488e-15 .. 4.338765e-13 | 4.877813e-17 .. 1.543351e-13 | 2.513018e-05 .. 0.04296092 | 2564 .. 2564 | 721.4456 | 7 | 10 | 5 | 5 | 19.02243 | -19.50066 | 723.0333 .. 722.767 | 0.6431202 |
| 82 | 1024 | 2468.640092303987 .. 2510.167438546871 | -0.0003368225 .. -0.004896033 | 0.0008588951 .. 0.0003851089 | -0.001195718 .. -0.005281142 | -2.433021e-15 .. -2.118669e-14 | 5.23886e-17 .. 9.378469e-14 | 2.574658e-05 .. 0.02337742 | 2564 .. 2564 | 759.4907 | 7 | 10 | 5 | 5 | 18.55963 | -19.02243 | 760.7409 .. 761.0948 | 0.6802197 |
| 83 | 1024 | 2438.870300692775 .. 2479.900734218312 | -0.0003547371 .. 0.009568594 | 0.0008357177 .. 0.0003162371 | -0.001190455 .. 0.009252357 | -2.507119e-15 .. 4.131679e-14 | 5.523122e-17 .. 1.179408e-13 | 2.622543e-05 .. 0.02641129 | 2564 .. 2564 | 796.5957 | 7 | 10 | 5 | 5 | 18.11187 | -18.55963 | 797.6779 .. 798.4939 | 0.6858748 |
| 84 | 1024 | 2409.809495372872 .. 2450.353954398244 | -0.008670044 .. -0.0282471 | 0.0007384224 .. 0.0002916735 | -0.009408467 .. -0.02853877 | -2.10243e-14 .. -1.404878e-13 | 2.889771e-16 .. 8.838024e-14 | 0.0001293185 .. 0.01795361 | 2580 .. 2564 | 832.8113 | 7 | 10 | 5 | 5 | 17.6788 | -18.11187 | 833.6879 .. 835.0538 | 0.6773117 |
| 85 | 1024 | 2381.432648558161 .. 2421.501716060622 | -0.0003633348 .. 0.01136095 | 0.0006555443 .. 0.0002394961 | -0.001018879 .. 0.01112145 | -2.435009e-15 .. 5.788485e-14 | 6.317231e-17 .. 1.382408e-13 | 2.643315e-05 .. 0.02656028 | 2564 .. 2564 | 868.1069 | 7 | 10 | 5 | 5 | 17.26004 | -17.6788 | 868.6306 .. 870.58 | 0.5923263 |
| 86 | 1024 | 2353.715896742346 .. 2393.319815671447 | -0.0008927707 .. -0.0007723463 | 0.0005372268 .. 0.0001914641 | -0.001429998 .. -0.0009638104 | -3.619407e-15 .. -5.706289e-15 | 1.054538e-16 .. 1.443463e-13 | 4.166393e-05 .. 0.02438055 | 2564 .. 2564 | 902.6914 | 7 | 10 | 5 | 5 | 16.85524 | -17.26004 | 903.117 .. 905.5424 | 0.6096405 |
| 87 | 1024 | 2326.636473776193 .. 2365.785161460157 | -0.000400295 .. -0.03696871 | 0.0004189655 .. 0.000183437 | -0.0008192605 .. -0.03715215 | -2.211804e-15 .. -2.487948e-13 | 7.499922e-17 .. 9.905844e-14 | 2.778e-05 .. 0.01479224 | 2628 .. 2564 | 936.4827 | 7 | 10 | 5 | 5 | 16.46401 | -16.85524 | 936.3604 .. 939.4835 | 0.618917 |
| 88 | 1024 | 2300.172648507885 .. 2338.875710305259 | -0.0003517445 .. -0.02851629 | 0.0003215419 .. 0.000134591 | -0.0006732864 .. -0.02865088 | -2.019401e-15 .. -1.999815e-13 | 7.836038e-17 .. 1.205028e-13 | 2.612605e-05 .. 0.01726415 | 2628 .. 2564 | 969.4632 | 7 | 10 | 5 | 5 | 16.08601 | -16.46401 | 969.1396 .. 972.7735 | 0.5758032 |
| 89 | 1024 | 2274.303666628806 .. 2312.570408871539 | -0.0004099174 .. -0.03550519 | 7.699389e-05 .. 9.6526e-05 | -0.0004869113 .. -0.03560172 | -1.739565e-15 .. -2.858723e-13 | 1.012215e-16 .. 1.178059e-13 | 2.833231e-05 .. 0.01467121 | 2628 .. 2564 | 1001.779 | 7 | 10 | 5 | 5 | 15.72085 | -16.08601 | 1001.029 .. 1005.438 | 0.6011575 |
Now we can plot the spectrum of all spectral orders in one plot, with each spectral order having a distinct color:
# plot the spectrum; the color of each SPORDER is specified through a matplotlib built-in colormap
matplotlib.rcParams['figure.figsize'] = (20, 7)
for i in range(len(echelle_data)):
plt.plot(echelle_data[i]['WAVELENGTH'], echelle_data[i]['FLUX'], color=plt.get_cmap('prism')(i/len(echelle_data)), label=echelle_data[i]['SPORDER'], alpha=0.7)
plt.xlabel('Wavelength [' + chr(197) + ']')
plt.ylabel("Flux [ergs/s/cm$^2$/" + chr(197) + "]")
plt.legend(loc='best')
plt.title("STIS Echelle Mode Spectrum")
plt.show()
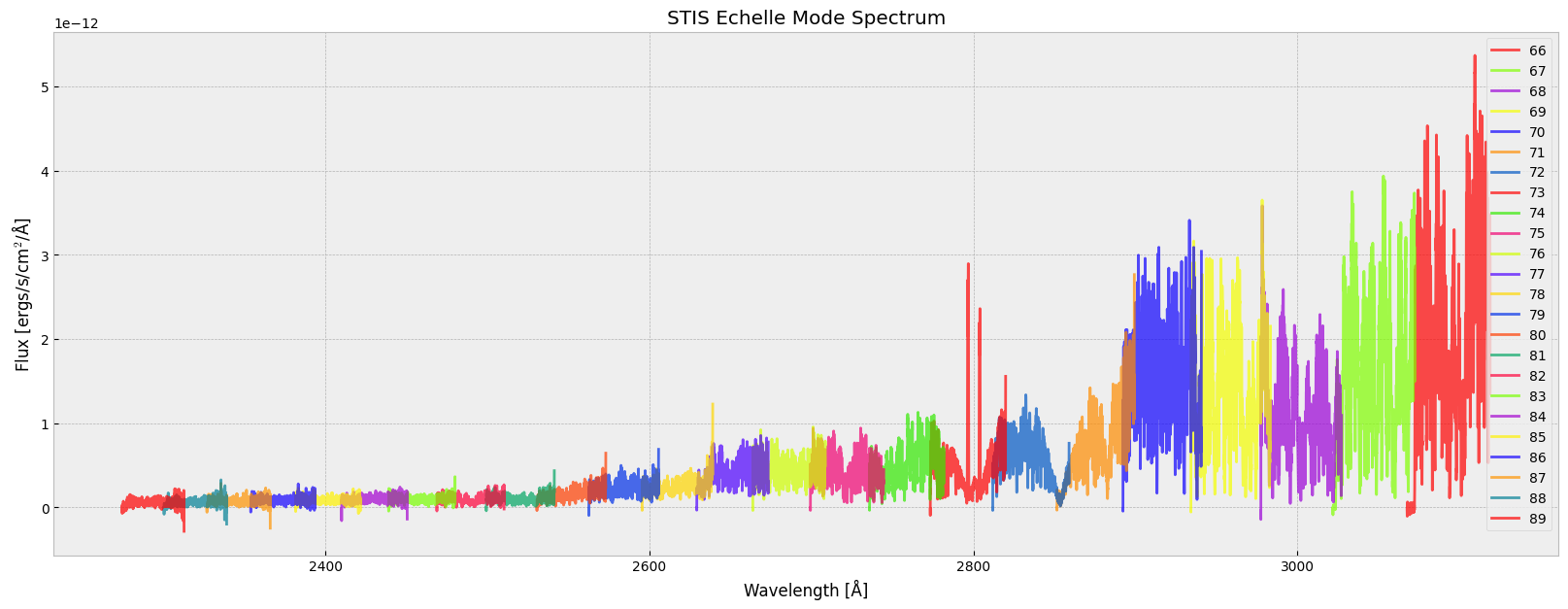
As the spectrum illustrates, each spectral order covers a specific wavelength range. Notice that some of the wavelength ranges overlap.
About this Notebook#
Author: Keyi Ding
Updated On: 2025-11-05
This tutorial was generated to be in compliance with the STScI style guides and would like to cite the Jupyter guide in particular.
Citations#
If you use astropy, matplotlib, astroquery, or numpy for published research, please cite the
authors. Follow these links for more information about citations:


