
Manual Recalibration with calwf3: Turning off the IR Linear Ramp Fit#
Learning Goals#
This notebook shows two reprocessing examples for WFC3/IR observations impacted by time-variable background (TVB).
By the end of this tutorial, you will:
Analyze exposure statistics for each read in an IMA file using
pstat.Reprocess a single exposure and an image association using
calwf3.Combine the reprocessed exposures using
astrodrizzle.
Table of Contents#
Introduction
1. Imports
2. Download the data
3. Query CRDS for reference files
4. Diagnose TVB and reprocess a single exposure
5. Reprocess multiple exposures in an association
6. Conclusions
Additional Resources
About the Notebook
Citations
Introduction#
Exposures in the F105W and F110W filters may be impacted by Helium I emission from the Earth’s atmosphere at 1.083 microns. This typically affects the reads taken closest in time to Earth occultation. The emission produces a flat background signal which is added to the total background in a subset of reads. In some cases, this non-linear signal may be strong enough to compromise the ramp fitting performed by calwf3, which is designed to flag and remove cosmic rays and saturated reads. The affected calibrated FLT data products will have much larger noise and a non-gaussian sky background.
This notebook demonstrates how to diagnose and correct for a non-linear background and is based on the ‘Last-minus-first’ technique described in WFC3 ISR 2016-16: Reprocessing WFC3/IR Exposures Affected by Time-Variable Backgrounds. This turns off the ramp fitting step in calwf3 and treats the IR detector like a CCD that accumulates charge and is read out only at the end of the exposure. In this case, the observed count rate is determined by simply subtracting the first from the last read of the detector and dividing by the time elapsed between the two reads.
While non-linear background also impacts the IR grisms, the method described here should not be used to correct G102 and G141 observations, which are affected by a combination of Helium I, Zodiacal background, and scattered Earth light, each of which varies spatially across the detector. More detail on correcting grism data is provided in WFC3 ISR 2020-04: The dispersed infrared background in WFC3 G102 and G141 observations.
1. Imports#
This notebook assumes you have installed the required libraries as described here.
We import:
glob for finding lists of files
os for setting environment variables
shutil for managing directories
matplotlib.pyplot for plotting data
astropy.io fits for accessing FITS files
astroquery.mast Observations for downloading data from MAST
ccdproc for building the association
drizzlepac astrodrizzle for combining images
stwcs for updating the World Coordinate System
wfc3tools calwf3 and pstat for calibrating WFC3 data and plotting statistics of WFC3 data
%matplotlib inline
import glob
import os
import shutil
import matplotlib.pyplot as plt
from astropy.io import fits
from astroquery.mast import Observations
from ccdproc import ImageFileCollection
from drizzlepac import astrodrizzle
from stwcs import updatewcs
from wfc3tools import calwf3, pstat
from IPython.display import clear_output
2. Download the data#
The following commands query MAST for the necessary products and then downloads them to the current directory. Here we obtain WFC3/IR observations from CANDELS program 12242, Visit BF. The data products requested are the ASN, RAW, IMA, FLT, and DRZ files.
Warning: this cell may take a few minutes to complete.#
data_list = Observations.query_criteria(obs_id='IBOHBF040')
Observations.download_products(
data_list['obsid'],
project='CALWF3',
download_dir='./data',
mrp_only=False,
productSubGroupDescription=['ASN', 'RAW', 'IMA', 'FLT', 'DRZ'])
science_files = glob.glob('data/mastDownload/HST/*/*fits')
for im in science_files:
root = os.path.basename(im)
new_path = os.path.join('.', root)
os.rename(im, new_path)
data_directory = './data'
try:
if os.path.isdir(data_directory):
shutil.rmtree(data_directory)
except Exception as e:
print(f"An error occured while deleting the directory {data_directory}: {e}")
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb7q_ima.fits to ./data/mastDownload/HST/ibohbfb7q/ibohbfb7q_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb7q_flt.fits to ./data/mastDownload/HST/ibohbfb7q/ibohbfb7q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb7q_raw.fits to ./data/mastDownload/HST/ibohbfb7q/ibohbfb7q_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb9q_ima.fits to ./data/mastDownload/HST/ibohbfb9q/ibohbfb9q_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb9q_flt.fits to ./data/mastDownload/HST/ibohbfb9q/ibohbfb9q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfb9q_raw.fits to ./data/mastDownload/HST/ibohbfb9q/ibohbfb9q_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbdq_ima.fits to ./data/mastDownload/HST/ibohbfbdq/ibohbfbdq_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbdq_flt.fits to ./data/mastDownload/HST/ibohbfbdq/ibohbfbdq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbdq_raw.fits to ./data/mastDownload/HST/ibohbfbdq/ibohbfbdq_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbgq_ima.fits to ./data/mastDownload/HST/ibohbfbgq/ibohbfbgq_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbgq_flt.fits to ./data/mastDownload/HST/ibohbfbgq/ibohbfbgq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbgq_raw.fits to ./data/mastDownload/HST/ibohbfbgq/ibohbfbgq_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbkq_ima.fits to ./data/mastDownload/HST/ibohbfbkq/ibohbfbkq_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbkq_flt.fits to ./data/mastDownload/HST/ibohbfbkq/ibohbfbkq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbkq_raw.fits to ./data/mastDownload/HST/ibohbfbkq/ibohbfbkq_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbpq_ima.fits to ./data/mastDownload/HST/ibohbfbpq/ibohbfbpq_ima.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbpq_flt.fits to ./data/mastDownload/HST/ibohbfbpq/ibohbfbpq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbfbpq_raw.fits to ./data/mastDownload/HST/ibohbfbpq/ibohbfbpq_raw.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbf040_asn.fits to ./data/mastDownload/HST/ibohbf040/ibohbf040_asn.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibohbf040_drz.fits to ./data/mastDownload/HST/ibohbf040/ibohbf040_drz.fits ...
[Done]
The association file for visit BF comprises six consecutive exposures in F105W acquired in a single visit over 3 orbits. Each orbit consists of two 1600 sec exposures, followed by the Earth occultation. Each exposure is dithered by a small fraction of the field of view, where the POSTARG values listed below are in arseconds.
image_collection = ImageFileCollection(
'./',
keywords=[
"asn_id",
"targname",
"filter",
"samp_seq",
"nsamp",
"exptime",
"postarg1",
"postarg2",
"date-obs",
"time-obs",
],
glob_include="*flt.fits",
ext=0,
)
try:
summary_table = image_collection.summary
if summary_table:
print(summary_table)
else:
print("No FITS files matched the pattern or no relevant data found.")
except Exception as e:
print(f"An error occurred while creating the summary table: {e}")
file asn_id targname ... postarg2 date-obs time-obs
------------------ --------- ------------ ... -------- ---------- --------
ibohbfb7q_flt.fits IBOHBF040 GOODN-N1-VBF ... -0.212 2013-01-06 18:23:01
ibohbfb9q_flt.fits IBOHBF040 GOODN-N1-VBF ... -0.03 2013-01-06 18:50:42
ibohbfbdq_flt.fits IBOHBF040 GOODN-N1-VBF ... 0.273 2013-01-06 19:18:23
ibohbfbgq_flt.fits IBOHBF040 GOODN-N1-VBF ... 0.091 2013-01-06 19:48:08
ibohbfbkq_flt.fits IBOHBF040 GOODN-N1-VBF ... 0.212 2013-01-06 20:15:49
ibohbfbpq_flt.fits IBOHBF040 GOODN-N1-VBF ... 0.394 2013-01-06 20:43:30
3. Query CRDS for reference files#
Before running calwf3, we need to set some environment variables for several subsequent calibration tasks.
We will point to a subdirectory called crds_cache/ using the IREF environment variable. The IREF variable is used for WFC3 reference files. Other instruments use other variables, e.g., JREF for ACS.
os.environ['CRDS_SERVER_URL'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_SERVER'] = 'https://hst-crds.stsci.edu'
os.environ['CRDS_PATH'] = './crds_cache'
os.environ['iref'] = './crds_cache/references/hst/wfc3/'
The code block below will query CRDS for the best reference files currently available for these datasets and update the header keywords to point to these new files. We will use the Python package os to run terminal commands. In the terminal, the line would be:
crds bestrefs --files [filename] --sync-references=1 --update-bestrefs
…where ‘filename’ is the name of your fits file.
Warning: this cell may take a few minutes to complete.#
raw_files = glob.glob('*_raw.fits')
for file in raw_files:
!crds bestrefs --files {file} --sync-references=1 --update-bestrefs
clear_output()
4. Diagnose TVB and reprocess a single exposure#
In this example, we assume that the observer desires to reprocess only a single exposure with the ramp fitting step turned off. This is done by setting the CRCORR switch to OMIT from the default value (PERFORM).
First, we list the contents of the image association ‘ibohbf040’. This provides the rootnames of the six dithered exposures.
fits.getdata('ibohbf040_asn.fits', 1)
FITS_rec([('IBOHBFB7Q', 'EXP-DTH', True), ('IBOHBFB9Q', 'EXP-DTH', True),
('IBOHBFBDQ', 'EXP-DTH', True), ('IBOHBFBGQ', 'EXP-DTH', True),
('IBOHBFBKQ', 'EXP-DTH', True), ('IBOHBFBPQ', 'EXP-DTH', True),
('IBOHBF040', 'PROD-DTH', True)],
dtype=(numpy.record, [('MEMNAME', 'S14'), ('MEMTYPE', 'S14'), ('MEMPRSNT', 'i1')]))
Here, we compare the first two images in the set: ‘ibohbfb7q’ and ‘ibohbfb9q’. The first is has a nominal background with a constant rate and the second has a strongly non-linear background.
b7q_data = fits.getdata('ibohbfb7q_flt.fits', ext=1)
b9q_data = fits.getdata('ibohbfb9q_flt.fits', ext=1)
fig = plt.figure(figsize=(15, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(b7q_data, vmin=0.25, vmax=1.25, cmap='Greys_r', origin='lower')
ax2.imshow(b9q_data, vmin=1.25, vmax=2.25, cmap='Greys_r', origin='lower')
ax1.set_title('ibohbfb7q (Linear Bkg)', fontsize=20)
ax2.set_title('ibohbfb9q (Non-linear Bkg)', fontsize=20)
Text(0.5, 1.0, 'ibohbfb9q (Non-linear Bkg)')
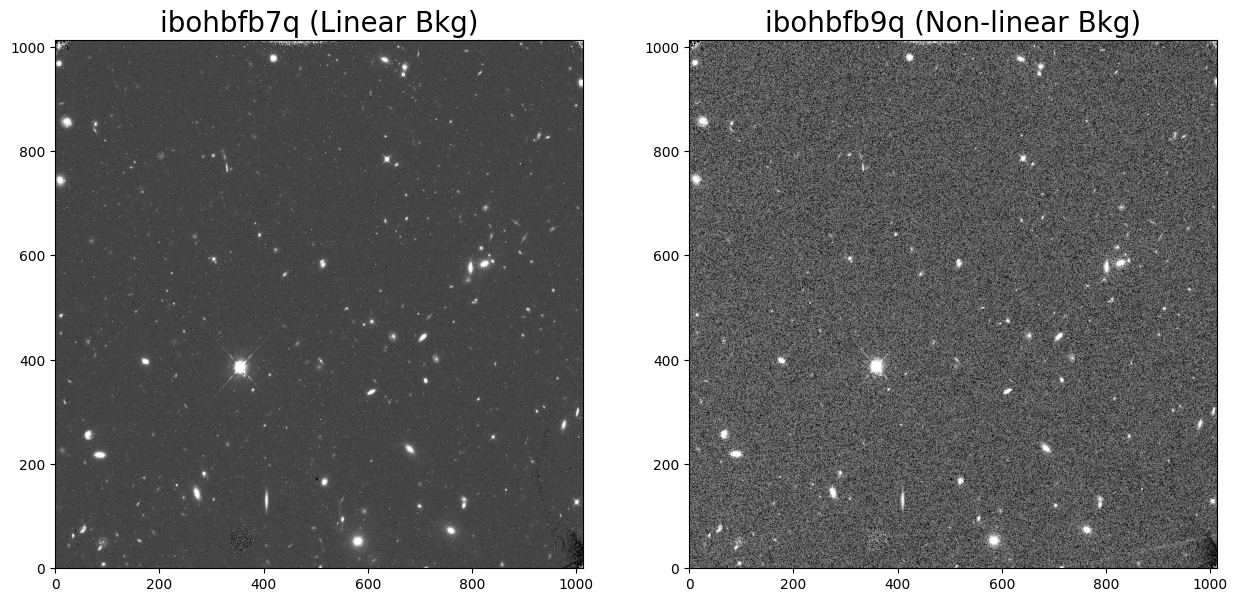
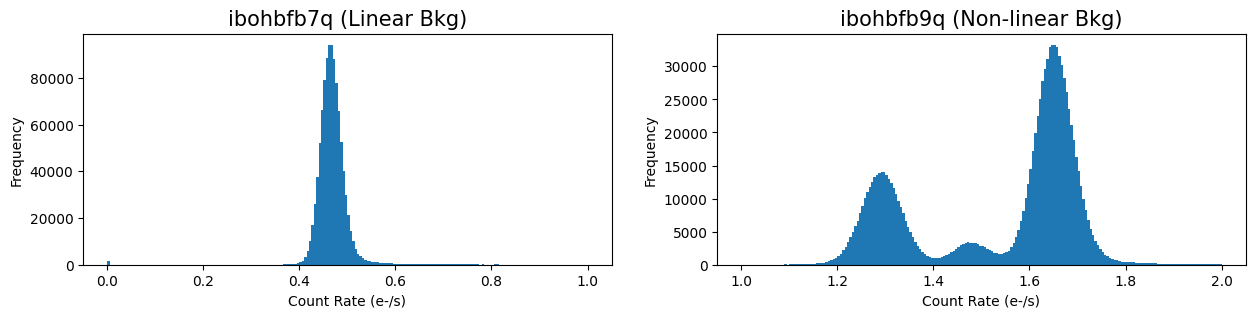
Next, we compare histograms of the two FLT frames. Exposure ‘ibohbfb9q’ has a strongly non-gaussian background with three separate peaks due to a poor ramp fit during calwf3 processing.
fig = plt.figure(figsize=(15, 3))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
n, bins, patches = ax1.hist(b7q_data.flatten(), bins=200, range=(0, 1))
n, bins, patches = ax2.hist(b9q_data.flatten(), bins=200, range=(1, 2))
ax1.set_title('ibohbfb7q (Linear Bkg)', fontsize=15)
ax1.set_xlabel('Count Rate (e-/s)')
ax1.set_ylabel('Frequency')
ax2.set_title('ibohbfb9q (Non-linear Bkg)', fontsize=15)
ax2.set_xlabel('Count Rate (e-/s)')
ax2.set_ylabel('Frequency')
Text(0, 0.5, 'Frequency')
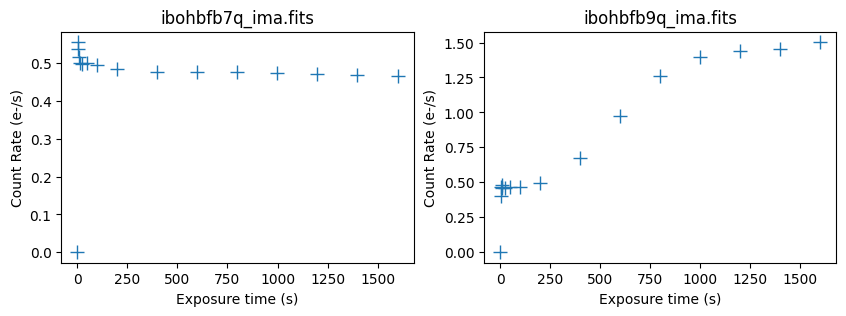
Next, we use pstat in wfc3tools to plot statistics for the individual reads in each IMA file.
Here, we plot the midpoint of each read in units of count rate. For the first image, the background is relatively constant throughout the exposure at 0.5 e/s. In the second image, the background quickly increases from a value of 0.5 e/s and levels off at ~1.5 e/s toward the end of the exposure.
imafiles = ('ibohbfb7q_ima.fits', 'ibohbfb9q_ima.fits')
fig, axarr = plt.subplots(1, 2)
axarr = axarr.reshape(-1)
fig.set_size_inches(10, 3)
fig.set_dpi(100)
for i, ima in enumerate(imafiles):
time, counts = pstat(ima, stat='midpt', units='rate', plot=False)
axarr[i].plot(time, counts, '+', markersize=10)
axarr[i].set_title(ima)
axarr[i].set_xlabel('Exposure time (s)')
axarr[i].set_ylabel('Count Rate (e-/s)')
To reprocess this image, we set the value of the header keyword CRCORR to “OMIT”. This will perform all steps in the calibration pipeline except for the ramp fitting. To see the current value of CRCORR, we use astropy.io.fits.getval( ) .
fits.getval('ibohbfb9q_raw.fits', 'CRCORR', 0)
'PERFORM'
Next, we edit the primary image header of the raw file to reflect the new value of CRCORR.
fits.setval('ibohbfb9q_raw.fits', 'CRCORR', value='OMIT')
Before running calwf3, we move the original pipeline products to a directory called orig/.
os.makedirs('orig/', exist_ok=True)
for file_pattern in ['ibohbf*_ima.fits', 'ibohbf*_flt.fits', 'ibohbf*_drz.fits']:
for file in glob.glob(file_pattern):
destination_path = os.path.join('orig', os.path.basename(file))
if os.path.isfile(destination_path):
os.remove(destination_path)
shutil.move(file, destination_path)
Finally, we run calwf3 on the single raw exposure.
calwf3('ibohbfb9q_raw.fits')
git tag: 0090c701-dirty
git branch: HEAD
HEAD @: 0090c701d894003cfc690e9f8d5fde81e6939090
CALBEG*** CALWF3 -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:03 UTC
Input ibohbfb9q_raw.fits
loading asn
LoadAsn: Processing SINGLE exposure
Trying to open ibohbfb9q_raw.fits...
Read in Primary header from ibohbfb9q_raw.fits...
Revising existing trailer file `ibohbfb9q.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:03 UTC
Input ibohbfb9q_raw.fits
Output ibohbfb9q_flt.fits
Trying to open ibohbfb9q_raw.fits...
Read in Primary header from ibohbfb9q_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfb9q_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1349 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1354 saturated pixels in imset 15
NLINCORR detected 1381 saturated pixels in imset 14
NLINCORR detected 1388 saturated pixels in imset 13
NLINCORR detected 1395 saturated pixels in imset 12
NLINCORR detected 1413 saturated pixels in imset 11
NLINCORR detected 1433 saturated pixels in imset 10
NLINCORR detected 1466 saturated pixels in imset 9
NLINCORR detected 1502 saturated pixels in imset 8
NLINCORR detected 1561 saturated pixels in imset 7
NLINCORR detected 1671 saturated pixels in imset 6
NLINCORR detected 2295 saturated pixels in imset 5
NLINCORR detected 2916 saturated pixels in imset 4
NLINCORR detected 3206 saturated pixels in imset 3
NLINCORR detected 3328 saturated pixels in imset 2
NLINCORR detected 3400 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.7852 as parnames[0]
==> Value of PHOTFLAM = 3.0608668e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR OMIT
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfb9q_ima.fits
Writing final image to ibohbfb9q_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:07 UTC
*** WF3IR complete ***
End 02-Dec-2025 20:20:07 UTC
*** CALWF3 complete ***
CALWF3 completion for ibohbfb9q_raw.fits
The product will be a single calibrated IMA and FLT image. We now compare the original FLT and the reprocessed FLT for a small 200x200 pixel region of the detector.
b9q_data = fits.getdata('orig/ibohbfb9q_flt.fits', ext=1)
b9q_newdata = fits.getdata('ibohbfb9q_flt.fits', ext=1)
fig = plt.figure(figsize=(15, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(b9q_data[520:720, 750:970], vmin=1.25, vmax=2.25, cmap='Greys_r', origin='lower')
ax2.imshow(b9q_newdata[520:720, 750:970], vmin=1.25, vmax=2.25, cmap='Greys_r', origin='lower')
ax1.set_title('ibohbfb9q (Original)', fontsize=20)
ax2.set_title('ibohbfb9q (Reprocessed)', fontsize=20)
Text(0.5, 1.0, 'ibohbfb9q (Reprocessed)')

Here we plot the image histogram showing the background in the original and reprocessed images.
fig = plt.figure(figsize=(15, 3))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
n, bins, patches = ax1.hist(b9q_data.flatten(), bins=200, range=(1, 2))
n, bins, patches = ax2.hist(b9q_newdata.flatten(), bins=200, range=(1, 2))
ax1.set_title('ibohbfb9q (Original FLT)', fontsize=15)
ax1.set_xlabel('Count Rate (e-/s)')
ax1.set_ylabel('Frequency')
ax2.set_title('ibohbfb9q (Reprocessed FLT)', fontsize=15)
ax2.set_xlabel('Count Rate (e-/s)')
ax2.set_ylabel('Frequency')
Text(0, 0.5, 'Frequency')
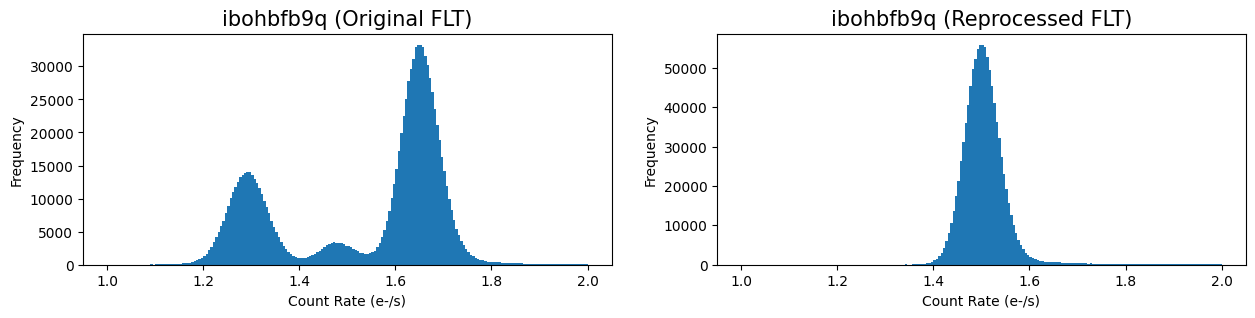
The non-gaussian image histogram is now corrected in the reprocessed FLT and the distribution is centered at a mean background of 1.5 e/s. One caveat of this approach is that cosmic-rays are not cleaned in the reprocessed image and will need to be corrected when combining the six FLT frames with AstroDrizzle. This is demonstrated in the next example.
5. Reprocess multiple exposures in an association#
In this example, we inspect the other images in the association to determine which are impacted by time-variable background, and we reprocess all six images with calwf3 and astrodrizzle.
Again, we list the contents of the association (asn) table.
dat = fits.getdata('ibohbf040_asn.fits', 1)
dat
FITS_rec([('IBOHBFB7Q', 'EXP-DTH', True), ('IBOHBFB9Q', 'EXP-DTH', True),
('IBOHBFBDQ', 'EXP-DTH', True), ('IBOHBFBGQ', 'EXP-DTH', True),
('IBOHBFBKQ', 'EXP-DTH', True), ('IBOHBFBPQ', 'EXP-DTH', True),
('IBOHBF040', 'PROD-DTH', True)],
dtype=(numpy.record, [('MEMNAME', 'S14'), ('MEMTYPE', 'S14'), ('MEMPRSNT', 'i1')]))
We can also print only the rootnames (ipppssoots) in the association.
dat['MEMNAME']
chararray(['IBOHBFB7Q', 'IBOHBFB9Q', 'IBOHBFBDQ', 'IBOHBFBGQ',
'IBOHBFBKQ', 'IBOHBFBPQ', 'IBOHBF040'], dtype='<U14')
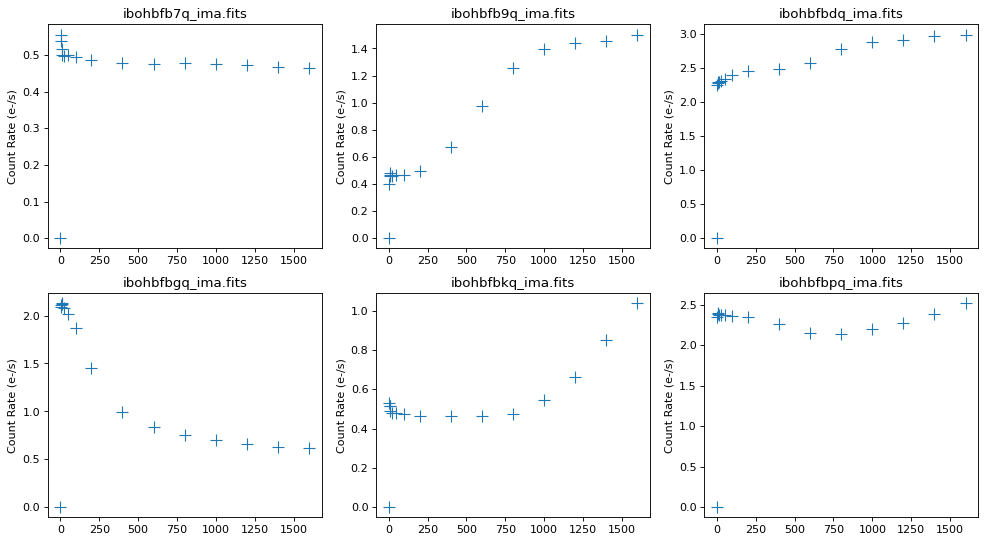
Using pstat, we can identify which of the six images are impacted by time-variable background.
Individual exposures b9q, bgq, and bkq show signs of strong time-variable background, where the change is more than a factor of 2. We will turn off the ramp fitting for these images and rerun calwf3.
imafiles = sorted(glob.glob('orig/*ima.fits'))
fig, axarr = plt.subplots(2, 3)
axarr = axarr.reshape(-1)
fig.set_size_inches(15, 8)
fig.set_dpi(80)
for i, ima in enumerate(imafiles):
time, counts = pstat(ima, stat='midpt', units='rate', plot=False)
axarr[i].plot(time, counts, '+', markersize=10)
axarr[i].set_title(ima[5:], fontsize=12)
axarr[i].set_ylabel('Count Rate (e-/s)')
Here we edit the primary image header of the three RAW images to set CRCORR to the value OMIT.
for rawfile in ['ibohbfb9q_raw.fits', 'ibohbfbgq_raw.fits', 'ibohbfbkq_raw.fits']:
fits.setval(rawfile, 'CRCORR', value='OMIT')
Next, we remove the calibrated products from the first example and then run calwf3 on the image association.
os.remove('ibohbfb9q_ima.fits')
os.remove('ibohbfb9q_flt.fits')
calwf3('ibohbf040_asn.fits')
# Alternatively, calwf3 may be run on a list of RAW files rather than the ASN
# for raws in glob.glob('ibohbf*_raw.fits'):
# calwf3(raws)
git tag: 0090c701-dirty
git branch: HEAD
HEAD @: 0090c701d894003cfc690e9f8d5fde81e6939090
CALBEG*** CALWF3 -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:10 UTC
Input ibohbf040_asn.fits
loading asn
LoadAsn: Processing FULL Association
Trying to open ibohbf040_asn.fits...
Read in Primary header from ibohbf040_asn.fits...
Revising existing trailer file `ibohbfb7q.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:10 UTC
Input ibohbfb7q_raw.fits
Output ibohbfb7q_flt.fits
Trying to open ibohbfb7q_raw.fits...
Read in Primary header from ibohbfb7q_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfb7q_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1348 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1351 saturated pixels in imset 15
NLINCORR detected 1377 saturated pixels in imset 14
NLINCORR detected 1382 saturated pixels in imset 13
NLINCORR detected 1389 saturated pixels in imset 12
NLINCORR detected 1408 saturated pixels in imset 11
NLINCORR detected 1424 saturated pixels in imset 10
NLINCORR detected 1458 saturated pixels in imset 9
NLINCORR detected 1498 saturated pixels in imset 8
NLINCORR detected 1548 saturated pixels in imset 7
NLINCORR detected 1601 saturated pixels in imset 6
NLINCORR detected 1648 saturated pixels in imset 5
NLINCORR detected 1685 saturated pixels in imset 4
NLINCORR detected 1737 saturated pixels in imset 3
NLINCORR detected 1799 saturated pixels in imset 2
NLINCORR detected 1859 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.7660 as parnames[0]
==> Value of PHOTFLAM = 3.0608666e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR PERFORM
CRREJTAB iref$u6a1748ri_crr.fits
CRIDCALC using 4 sigma rejection threshold
256 bad DQ mask
4 max CRs for UNSTABLE
88 pixels detected as unstable
CRCORR COMPLETE
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfb7q_ima.fits
Writing final image to ibohbfb7q_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:15 UTC
*** WF3IR complete ***
RPTCORR OMIT
Revising existing trailer file `ibohbfb9q.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:15 UTC
Input ibohbfb9q_raw.fits
Output ibohbfb9q_flt.fits
Trying to open ibohbfb9q_raw.fits...
Read in Primary header from ibohbfb9q_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfb9q_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1349 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1354 saturated pixels in imset 15
NLINCORR detected 1381 saturated pixels in imset 14
NLINCORR detected 1388 saturated pixels in imset 13
NLINCORR detected 1395 saturated pixels in imset 12
NLINCORR detected 1413 saturated pixels in imset 11
NLINCORR detected 1433 saturated pixels in imset 10
NLINCORR detected 1466 saturated pixels in imset 9
NLINCORR detected 1502 saturated pixels in imset 8
NLINCORR detected 1561 saturated pixels in imset 7
NLINCORR detected 1671 saturated pixels in imset 6
NLINCORR detected 2295 saturated pixels in imset 5
NLINCORR detected 2916 saturated pixels in imset 4
NLINCORR detected 3206 saturated pixels in imset 3
NLINCORR detected 3328 saturated pixels in imset 2
NLINCORR detected 3400 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.7852 as parnames[0]
==> Value of PHOTFLAM = 3.0608668e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR OMIT
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfb9q_ima.fits
Writing final image to ibohbfb9q_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:18 UTC
*** WF3IR complete ***
RPTCORR OMIT
Revising existing trailer file `ibohbfbdq.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:18 UTC
Input ibohbfbdq_raw.fits
Output ibohbfbdq_flt.fits
Trying to open ibohbfbdq_raw.fits...
Read in Primary header from ibohbfbdq_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfbdq_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1347 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1350 saturated pixels in imset 15
NLINCORR detected 1377 saturated pixels in imset 14
NLINCORR detected 1386 saturated pixels in imset 13
NLINCORR detected 1391 saturated pixels in imset 12
NLINCORR detected 1419 saturated pixels in imset 11
NLINCORR detected 1436 saturated pixels in imset 10
NLINCORR detected 1474 saturated pixels in imset 9
NLINCORR detected 1554 saturated pixels in imset 8
NLINCORR detected 2226 saturated pixels in imset 7
NLINCORR detected 3004 saturated pixels in imset 6
NLINCORR detected 3323 saturated pixels in imset 5
NLINCORR detected 3371 saturated pixels in imset 4
NLINCORR detected 3396 saturated pixels in imset 3
NLINCORR detected 3413 saturated pixels in imset 2
NLINCORR detected 3434 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.8044 as parnames[0]
==> Value of PHOTFLAM = 3.060867e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR PERFORM
CRREJTAB iref$u6a1748ri_crr.fits
CRIDCALC using 4 sigma rejection threshold
256 bad DQ mask
4 max CRs for UNSTABLE
53 pixels detected as unstable
CRCORR COMPLETE
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfbdq_ima.fits
Writing final image to ibohbfbdq_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:23 UTC
*** WF3IR complete ***
RPTCORR OMIT
Revising existing trailer file `ibohbfbgq.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:23 UTC
Input ibohbfbgq_raw.fits
Output ibohbfbgq_flt.fits
Trying to open ibohbfbgq_raw.fits...
Read in Primary header from ibohbfbgq_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfbgq_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1345 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1347 saturated pixels in imset 15
NLINCORR detected 1375 saturated pixels in imset 14
NLINCORR detected 1381 saturated pixels in imset 13
NLINCORR detected 1387 saturated pixels in imset 12
NLINCORR detected 1404 saturated pixels in imset 11
NLINCORR detected 1423 saturated pixels in imset 10
NLINCORR detected 1460 saturated pixels in imset 9
NLINCORR detected 1494 saturated pixels in imset 8
NLINCORR detected 1540 saturated pixels in imset 7
NLINCORR detected 1572 saturated pixels in imset 6
NLINCORR detected 1615 saturated pixels in imset 5
NLINCORR detected 1632 saturated pixels in imset 4
NLINCORR detected 1647 saturated pixels in imset 3
NLINCORR detected 1662 saturated pixels in imset 2
NLINCORR detected 1680 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.8251 as parnames[0]
==> Value of PHOTFLAM = 3.0608672e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR OMIT
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfbgq_ima.fits
Writing final image to ibohbfbgq_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:26 UTC
*** WF3IR complete ***
RPTCORR OMIT
Revising existing trailer file `ibohbfbkq.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:26 UTC
Input ibohbfbkq_raw.fits
Output ibohbfbkq_flt.fits
Trying to open ibohbfbkq_raw.fits...
Read in Primary header from ibohbfbkq_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfbkq_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1346 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1350 saturated pixels in imset 15
NLINCORR detected 1377 saturated pixels in imset 14
NLINCORR detected 1384 saturated pixels in imset 13
NLINCORR detected 1389 saturated pixels in imset 12
NLINCORR detected 1406 saturated pixels in imset 11
NLINCORR detected 1428 saturated pixels in imset 10
NLINCORR detected 1459 saturated pixels in imset 9
NLINCORR detected 1490 saturated pixels in imset 8
NLINCORR detected 1530 saturated pixels in imset 7
NLINCORR detected 1568 saturated pixels in imset 6
NLINCORR detected 1597 saturated pixels in imset 5
NLINCORR detected 1624 saturated pixels in imset 4
NLINCORR detected 1648 saturated pixels in imset 3
NLINCORR detected 1683 saturated pixels in imset 2
NLINCORR detected 1862 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.8443 as parnames[0]
==> Value of PHOTFLAM = 3.0608674e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR OMIT
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfbkq_ima.fits
Writing final image to ibohbfbkq_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:30 UTC
*** WF3IR complete ***
RPTCORR OMIT
Revising existing trailer file `ibohbfbpq.tra'.
CALBEG*** WF3IR -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:30 UTC
Input ibohbfbpq_raw.fits
Output ibohbfbpq_flt.fits
Trying to open ibohbfbpq_raw.fits...
Read in Primary header from ibohbfbpq_raw.fits...
APERTURE IR-FIX
FILTER F105W
DETECTOR IR
Reading data from ibohbfbpq_raw.fits ...
CCDTAB iref$t2c16200i_ccd.fits
CCDTAB PEDIGREE=Ground
CCDTAB DESCRIP =Reference data based on Thermal-Vac #3, gain=2.5 results for IR-4
CCDTAB DESCRIP =Readnoise,gain,saturation from TV3,MEB2 values. ISRs 2008-25,39,50
readnoise =20.2,19.8,19.9,20.1
gain =2.34,2.37,2.31,2.38
DQICORR PERFORM
DQITAB iref$3562033hi_bpx.fits
DQITAB PEDIGREE=INFLIGHT 01/11/2012 12/10/2013
DQITAB DESCRIP =Bad Pixel Table generated using Cycle 20 Flats and Darks-----------
DQICORR COMPLETE
ZSIGCORR PERFORM
ZSIGCORR detected 1285 saturated pixels in 0th read
ZSIGCORR detected 1347 saturated pixels in 1st read
ZSIGCORR COMPLETE
BLEVCORR PERFORM
OSCNTAB iref$q911321mi_osc.fits
OSCNTAB PEDIGREE=GROUND
OSCNTAB DESCRIP =Initial values for ground test data processing
BLEVCORR COMPLETE
ZOFFCORR PERFORM
ZOFFCORR COMPLETE
NOISCORR PERFORM
Uncertainty array initialized.
NOISCORR COMPLETE
NLINCORR PERFORM
NLINFILE iref$9au15283i_lin.fits
NLINFILE PEDIGREE=INFLIGHT 29/03/2011 25/11/2012
NLINFILE DESCRIP =Non-linearity, pixel-based correction from WFC3 on-orbit frames
NLINCORR detected 1285 saturated pixels in imset 16
NLINCORR detected 1351 saturated pixels in imset 15
NLINCORR detected 1378 saturated pixels in imset 14
NLINCORR detected 1382 saturated pixels in imset 13
NLINCORR detected 1390 saturated pixels in imset 12
NLINCORR detected 1409 saturated pixels in imset 11
NLINCORR detected 1426 saturated pixels in imset 10
NLINCORR detected 1463 saturated pixels in imset 9
NLINCORR detected 1507 saturated pixels in imset 8
NLINCORR detected 1634 saturated pixels in imset 7
NLINCORR detected 1980 saturated pixels in imset 6
NLINCORR detected 2560 saturated pixels in imset 5
NLINCORR detected 3029 saturated pixels in imset 4
NLINCORR detected 3278 saturated pixels in imset 3
NLINCORR detected 3359 saturated pixels in imset 2
NLINCORR detected 3392 saturated pixels in imset 1
NLINCORR COMPLETE
DARKCORR PERFORM
DARKFILE iref$35620245i_drk.fits
DARKFILE PEDIGREE=INFLIGHT 20/09/2009 15/11/2016
DARKFILE DESCRIP =Dark Created from 154 frames spanning cycles 17 to 24--------------
DARKCORR using dark imset 16 for imset 16 with exptime= 0
DARKCORR using dark imset 15 for imset 15 with exptime= 2.93229
DARKCORR using dark imset 14 for imset 14 with exptime= 5.86458
DARKCORR using dark imset 13 for imset 13 with exptime= 8.79687
DARKCORR using dark imset 12 for imset 12 with exptime= 11.7292
DARKCORR using dark imset 11 for imset 11 with exptime= 24.2297
DARKCORR using dark imset 10 for imset 10 with exptime= 49.2302
DARKCORR using dark imset 9 for imset 9 with exptime= 99.2306
DARKCORR using dark imset 8 for imset 8 with exptime= 199.231
DARKCORR using dark imset 7 for imset 7 with exptime= 399.231
DARKCORR using dark imset 6 for imset 6 with exptime= 599.232
DARKCORR using dark imset 5 for imset 5 with exptime= 799.232
DARKCORR using dark imset 4 for imset 4 with exptime= 999.232
DARKCORR using dark imset 3 for imset 3 with exptime= 1199.23
DARKCORR using dark imset 2 for imset 2 with exptime= 1399.23
DARKCORR using dark imset 1 for imset 1 with exptime= 1599.23
DARKCORR COMPLETE
PHOTCORR PERFORM
IMPHTTAB iref$8ch15233i_imp.fits
IMPHTTAB PEDIGREE=INFLIGHT 08/05/2009 01/09/2024
IMPHTTAB DESCRIP =Time-dependent image photometry reference table (IMPHTTAB)---------
Found parameterized variable 1.
NUMPAR=1, N=1
Allocated 1 parnames
Adding parameter mjd#56298.8635 as parnames[0]
==> Value of PHOTFLAM = 3.0608676e-20
==> Value of PHOTPLAM = 10551.047
==> Value of PHOTBW = 845.61804
PHOTCORR COMPLETE
UNITCORR PERFORM
UNITCORR COMPLETE
CRCORR PERFORM
CRREJTAB iref$u6a1748ri_crr.fits
CRIDCALC using 4 sigma rejection threshold
256 bad DQ mask
4 max CRs for UNSTABLE
64 pixels detected as unstable
CRCORR COMPLETE
FLATCORR PERFORM
PFLTFILE iref$4ac19225i_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
PFLTFILE DESCRIP =Sky Flat from Combined In-flight observations between 2009 and 2019
DFLTFILE iref$4ac18263i_dfl.fits
DFLTFILE PEDIGREE=INFLIGHT 31/07/2009 05/12/2019
DFLTFILE DESCRIP =Delta-Flat for IR Blobs by Date of Appearance from Sky Flat Dataset
FLATCORR COMPLETE
Writing calibrated readouts to ibohbfbpq_ima.fits
Writing final image to ibohbfbpq_flt.fits
with trimx = 5,5, trimy = 5,5
End 02-Dec-2025 20:20:35 UTC
*** WF3IR complete ***
RPTCORR OMIT
CALBEG*** WF3DTH -- Version 3.7.2 (Apr-15-2024) ***
Begin 02-Dec-2025 20:20:35 UTC
Astrodrizzle needs to be run in order to generate
a geometrically corrected, drizzle-combined product.
Warning Can't find input file "ibohbfb7q_spt.fits"
Warning Can't find input file "ibohbfb9q_spt.fits"
Warning Can't find input file "ibohbfbdq_spt.fits"
Warning Can't find input file "ibohbfbgq_spt.fits"
Warning Can't find input file "ibohbfbkq_spt.fits"
Warning Can't find input file "ibohbfbpq_spt.fits"
End 02-Dec-2025 20:20:35 UTC
*** WF3DTH complete ***
Trying to open ibohbf040_asn.fits...
Updated Global Header for ibohbf040_asn.fits...
End 02-Dec-2025 20:20:35 UTC
*** CALWF3 complete ***
CALWF3 completion for ibohbf040_asn.fits
Finally we combine the reprocessed FLTs with AstroDrizzle.
First, the World Coordinate System (WCS) of the calibrated images must be updated using updatewcs. This prepares the image for astrodrizzle to apply the various components of geometric distortion correction.
When the parameter use_db=False, the WCS will be based on the coordinates of the Guide Star Catalogs in use at the time. No realignment of the images is performed, and this typically gives the best ‘relative’ astrometry between exposures in a visit, either in the same filter or across multiple filters.
When use_db=True, the software will connect to the astrometry database and update the WCS to an absolute frame of reference, typically based on an external catalog such as Gaia. Here, the quality of the fit is dependent on the number of bright sources in each image, and in some cased the relative astrometry may not be optimal.
for flts in glob.glob('ibohbf*_flt.fits'):
updatewcs.updatewcs(input=flts, use_db=False)
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
- IDCTAB: Distortion model from row 1 for chip 1 : F105W
Next, we use AstroDrizzle to combine the FLT frames, making use of internal CR-flagging algorithms to clean the images.
astrodrizzle.AstroDrizzle(input='ibohbf040_asn.fits', mdriztab=True, preserve=False, clean=True)
INFO:drizzlepac.util:Setting up logfile : astrodrizzle.log
Setting up logfile : astrodrizzle.log
INFO:drizzlepac.astrodrizzle:AstroDrizzle log file: astrodrizzle.log
AstroDrizzle log file: astrodrizzle.log
INFO:drizzlepac.astrodrizzle:AstroDrizzle Version 3.10.0 started at: 20:20:35.953 (02/12/2025)
AstroDrizzle Version 3.10.0 started at: 20:20:35.953 (02/12/2025)
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:Version Information
INFO:drizzlepac.astrodrizzle:--------------------
INFO:drizzlepac.astrodrizzle:Python Version 3.11.14 | packaged by conda-forge | (main, Oct 22 2025, 22:46:25) [GCC 14.3.0]
INFO:drizzlepac.astrodrizzle:numpy Version -> 2.3.5
INFO:drizzlepac.astrodrizzle:astropy Version -> 7.2.0
INFO:drizzlepac.astrodrizzle:stwcs Version -> 1.7.5
INFO:drizzlepac.astrodrizzle:photutils Version -> 2.3.0
INFO:drizzlepac.util:==== Processing Step Initialization started at 20:20:35.959 (02/12/2025)
==== Processing Step Initialization started at 20:20:35.959 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.processInput:Executing serially
INFO:drizzlepac.processInput:Setting up output name: ibohbf040_drz.fits
INFO:drizzlepac.processInput:Reading in MDRIZTAB parameters for 6 files
Reading in MDRIZTAB parameters for 6 files
INFO:drizzlepac.mdzhandler:- MDRIZTAB: AstroDrizzle parameters read from row 3.
- MDRIZTAB: AstroDrizzle parameters read from row 3.
INFO:drizzlepac.processInput:-Creating imageObject List as input for processing steps.
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfb7q_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfb9q_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfbdq_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfbgq_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfbkq_flt.fits[DQ,1]
INFO:drizzlepac.resetbits:Reset bit values of 4096 to a value of 0 in ibohbfbpq_flt.fits[DQ,1]
INFO:drizzlepac.processInput:-Creating output WCS.
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 189.12062959697133 62.23566816080676
CRVAL : 189.12062959697133 62.23566816080676
INFO:astropy.wcs.wcs:CRPIX : 546.5 482.0
CRPIX : 546.5 482.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
INFO:astropy.wcs.wcs:NAXIS : 1093 964
NAXIS : 1093 964
INFO:drizzlepac.processInput:********************************************************************************
********************************************************************************
INFO:drizzlepac.processInput:*
*
INFO:drizzlepac.processInput:* Estimated memory usage: up to 79 Mb.
* Estimated memory usage: up to 79 Mb.
INFO:drizzlepac.processInput:* Output image size: 1093 X 964 pixels.
* Output image size: 1093 X 964 pixels.
INFO:drizzlepac.processInput:* Output image file: ~ 12 Mb.
* Output image file: ~ 12 Mb.
INFO:drizzlepac.processInput:* Cores available: 4
* Cores available: 4
INFO:drizzlepac.processInput:*
*
INFO:drizzlepac.processInput:********************************************************************************
********************************************************************************
INFO:drizzlepac.util:==== Processing Step Initialization finished at 20:20:36.644 (02/12/2025)
==== Processing Step Initialization finished at 20:20:36.644 (02/12/2025)
INFO:drizzlepac.astrodrizzle:USER INPUT PARAMETERS common to all Processing Steps:
INFO:drizzlepac.astrodrizzle: build : True
INFO:drizzlepac.astrodrizzle: coeffs : True
INFO:drizzlepac.astrodrizzle: context : True
INFO:drizzlepac.astrodrizzle: crbit : 4096
INFO:drizzlepac.astrodrizzle: group :
INFO:drizzlepac.astrodrizzle: in_memory : False
INFO:drizzlepac.astrodrizzle: input : ibohbf040_asn.fits
INFO:drizzlepac.astrodrizzle: mdriztab : True
INFO:drizzlepac.astrodrizzle: num_cores : None
INFO:drizzlepac.astrodrizzle: output :
INFO:drizzlepac.astrodrizzle: proc_unit : native
INFO:drizzlepac.astrodrizzle: resetbits : 4096
INFO:drizzlepac.astrodrizzle: rules_file :
INFO:drizzlepac.astrodrizzle: runfile :
INFO:drizzlepac.astrodrizzle: stepsize : 10
INFO:drizzlepac.astrodrizzle: updatewcs : False
INFO:drizzlepac.astrodrizzle: wcskey :
INFO:drizzlepac.util:==== Processing Step Static Mask started at 20:20:36.653 (02/12/2025)
==== Processing Step Static Mask started at 20:20:36.653 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.staticMask:USER INPUT PARAMETERS for Static Mask Step:
INFO:drizzlepac.staticMask: static : True
INFO:drizzlepac.staticMask: static_sig : 4.0
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 0.464975; rms = 0.024820; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.497733; rms = 0.038513; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 2.976076; rms = 0.053656; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 0.610324; rms = 0.027323; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 1.034977; rms = 0.033774; static_sig = 4.00
INFO:drizzlepac.staticMask:Computing static mask:
INFO:drizzlepac.staticMask: mode = 2.475992; rms = 0.054044; static_sig = 4.00
INFO:drizzlepac.staticMask:Saving static mask to disk: ./_1014x1014_1_staticMask.fits
INFO:drizzlepac.util:==== Processing Step Static Mask finished at 20:20:36.795 (02/12/2025)
==== Processing Step Static Mask finished at 20:20:36.795 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Subtract Sky started at 20:20:36.797 (02/12/2025)
==== Processing Step Subtract Sky started at 20:20:36.797 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.sky:USER INPUT PARAMETERS for Sky Subtraction Step:
INFO:drizzlepac.sky: sky_bits : 16
INFO:drizzlepac.sky: skyclip : 5
INFO:drizzlepac.sky: skyfile :
INFO:drizzlepac.sky: skylower : -100.0
INFO:drizzlepac.sky: skylsigma : 4.0
INFO:drizzlepac.sky: skymask_cat :
INFO:drizzlepac.sky: skymethod : match
INFO:drizzlepac.sky: skystat : mode
INFO:drizzlepac.sky: skysub : True
INFO:drizzlepac.sky: skyupper : None
INFO:drizzlepac.sky: skyuser :
INFO:drizzlepac.sky: skyusigma : 4.0
INFO:drizzlepac.sky: skywidth : 0.10000000149011612
INFO:drizzlepac.sky: use_static : True
INFO:stsci.skypac.utils:***** skymatch started on 2025-12-02 20:20:36.865207
***** skymatch started on 2025-12-02 20:20:36.865207
INFO:stsci.skypac.utils: Version 1.0.11
Version 1.0.11
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:'skymatch' task will apply computed sky differences to input image file(s).
'skymatch' task will apply computed sky differences to input image file(s).
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
INFO:stsci.skypac.utils:'MDRIZSKY' header keyword will represent sky value *computed* from data.
'MDRIZSKY' header keyword will represent sky value *computed* from data.
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- User specified keywords: -----
----- User specified keywords: -----
INFO:stsci.skypac.utils: Sky Value Keyword: 'MDRIZSKY'
Sky Value Keyword: 'MDRIZSKY'
INFO:stsci.skypac.utils: Data Units Keyword: 'BUNIT'
Data Units Keyword: 'BUNIT'
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Input file list: -----
----- Input file list: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfb7q_flt.fits'
** Input image: 'ibohbfb7q_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfb7q_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfb7q_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfb9q_flt.fits'
** Input image: 'ibohbfb9q_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfb9q_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfb9q_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfbdq_flt.fits'
** Input image: 'ibohbfbdq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfbdq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfbdq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfbgq_flt.fits'
** Input image: 'ibohbfbgq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfbgq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfbgq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfbkq_flt.fits'
** Input image: 'ibohbfbkq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfbkq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfbkq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: ** Input image: 'ibohbfbpq_flt.fits'
** Input image: 'ibohbfbpq_flt.fits'
INFO:stsci.skypac.utils: EXT: 'SCI',1; MASK: ibohbfbpq_skymatch_mask_sci1.fits[0]
EXT: 'SCI',1; MASK: ibohbfbpq_skymatch_mask_sci1.fits[0]
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Sky statistics parameters: -----
----- Sky statistics parameters: -----
INFO:stsci.skypac.utils: statistics function: 'mode'
statistics function: 'mode'
INFO:stsci.skypac.utils: lower = -100.0
lower = -100.0
INFO:stsci.skypac.utils: upper = None
upper = None
INFO:stsci.skypac.utils: nclip = 5
nclip = 5
INFO:stsci.skypac.utils: lsigma = 4.0
lsigma = 4.0
INFO:stsci.skypac.utils: usigma = 4.0
usigma = 4.0
INFO:stsci.skypac.utils: binwidth = 0.10000000149011612
binwidth = 0.10000000149011612
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Data->Brightness conversion parameters for input files: -----
----- Data->Brightness conversion parameters for input files: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfb7q_flt.fits
* Image: ibohbfb7q_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfb9q_flt.fits
* Image: ibohbfb9q_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfbdq_flt.fits
* Image: ibohbfbdq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfbgq_flt.fits
* Image: ibohbfbgq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfbkq_flt.fits
* Image: ibohbfbkq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image: ibohbfbpq_flt.fits
* Image: ibohbfbpq_flt.fits
INFO:stsci.skypac.utils: EXT = 'SCI',1
EXT = 'SCI',1
INFO:stsci.skypac.utils: Data units type: COUNT-RATE
Data units type: COUNT-RATE
INFO:stsci.skypac.utils: Conversion factor (data->brightness): 60.79743434827051
Conversion factor (data->brightness): 60.79743434827051
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:----- Computing differences in sky values in overlapping regions: -----
----- Computing differences in sky values in overlapping regions: -----
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfb7q_flt.fits['SCI',1]' SKY = 0 [brightness units]
* Image 'ibohbfb7q_flt.fits['SCI',1]' SKY = 0 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0
- EXT = 'SCI',1 delta(MDRIZSKY) = 0
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfb9q_flt.fits['SCI',1]' SKY = 62.8273 [brightness units]
* Image 'ibohbfb9q_flt.fits['SCI',1]' SKY = 62.8273 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 1.03339
- EXT = 'SCI',1 delta(MDRIZSKY) = 1.03339
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfbdq_flt.fits['SCI',1]' SKY = 152.745 [brightness units]
* Image 'ibohbfbdq_flt.fits['SCI',1]' SKY = 152.745 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 2.51235
- EXT = 'SCI',1 delta(MDRIZSKY) = 2.51235
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfbgq_flt.fits['SCI',1]' SKY = 8.83832 [brightness units]
* Image 'ibohbfbgq_flt.fits['SCI',1]' SKY = 8.83832 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0.145373
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.145373
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfbkq_flt.fits['SCI',1]' SKY = 34.7582 [brightness units]
* Image 'ibohbfbkq_flt.fits['SCI',1]' SKY = 34.7582 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 0.571706
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.571706
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils: * Image 'ibohbfbpq_flt.fits['SCI',1]' SKY = 122.268 [brightness units]
* Image 'ibohbfbpq_flt.fits['SCI',1]' SKY = 122.268 [brightness units]
INFO:stsci.skypac.utils: Updating sky of image extension(s) [data units]:
Updating sky of image extension(s) [data units]:
INFO:stsci.skypac.utils: - EXT = 'SCI',1 delta(MDRIZSKY) = 2.01108
- EXT = 'SCI',1 delta(MDRIZSKY) = 2.01108
INFO:stsci.skypac.utils:
INFO:stsci.skypac.utils:***** skymatch ended on 2025-12-02 20:20:39.038297
***** skymatch ended on 2025-12-02 20:20:39.038297
INFO:stsci.skypac.utils:TOTAL RUN TIME: 0:00:02.173090
TOTAL RUN TIME: 0:00:02.173090
INFO:drizzlepac.util:==== Processing Step Subtract Sky finished at 20:20:39.086 (02/12/2025)
==== Processing Step Subtract Sky finished at 20:20:39.086 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Separate Drizzle started at 20:20:39.088 (02/12/2025)
==== Processing Step Separate Drizzle started at 20:20:39.088 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.adrizzle:Interpreted paramDict with single=True as:
{'build': True, 'stepsize': 10, 'coeffs': True, 'wcskey': '', 'kernel': 'turbo', 'wt_scl': 'exptime', 'pixfrac': 1.0, 'fillval': 'INDEF', 'bits': 528, 'compress': False, 'units': 'cps'}
INFO:drizzlepac.adrizzle:USER INPUT PARAMETERS for Separate Drizzle Step:
INFO:drizzlepac.adrizzle: bits : 528
INFO:drizzlepac.adrizzle: build : False
INFO:drizzlepac.adrizzle: clean : True
INFO:drizzlepac.adrizzle: coeffs : True
INFO:drizzlepac.adrizzle: compress : False
INFO:drizzlepac.adrizzle: crbit : None
INFO:drizzlepac.adrizzle: fillval : INDEF
INFO:drizzlepac.adrizzle: kernel : turbo
INFO:drizzlepac.adrizzle: num_cores : None
INFO:drizzlepac.adrizzle: pixfrac : 1.0
INFO:drizzlepac.adrizzle: proc_unit : electrons
INFO:drizzlepac.adrizzle: rules_file : None
INFO:drizzlepac.adrizzle: stepsize : 10
INFO:drizzlepac.adrizzle: units : cps
INFO:drizzlepac.adrizzle: wcskey :
INFO:drizzlepac.adrizzle: wht_type : None
INFO:drizzlepac.adrizzle: wt_scl : exptime
INFO:drizzlepac.adrizzle: **Using sub-sampling value of 10 for kernel turbo
INFO:drizzlepac.adrizzle:Running Drizzle to create output frame with WCS of:
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 189.12062959697133 62.23566816080676
CRVAL : 189.12062959697133 62.23566816080676
INFO:astropy.wcs.wcs:CRPIX : 546.5 482.0
CRPIX : 546.5 482.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
INFO:astropy.wcs.wcs:NAXIS : 1093 964
NAXIS : 1093 964
INFO:drizzlepac.adrizzle:Executing 4 parallel workers
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.000000 to ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 1.033388 to ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 2.512355 to ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.adrizzle:Applying sky value of 0.145373 to ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfb7q_single_sci.fits
-Generating simple FITS output: ibohbfb7q_single_sci.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfb9q_single_sci.fits
-Generating simple FITS output: ibohbfb9q_single_sci.fits
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbdq_single_sci.fits
-Generating simple FITS output: ibohbfbdq_single_sci.fits
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbgq_single_sci.fits
-Generating simple FITS output: ibohbfbgq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb7q_single_sci.fits
Writing out image to disk: ibohbfb7q_single_sci.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb9q_single_sci.fits
Writing out image to disk: ibohbfb9q_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbdq_single_sci.fits
Writing out image to disk: ibohbfbdq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbgq_single_sci.fits
Writing out image to disk: ibohbfbgq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb7q_single_wht.fits
Writing out image to disk: ibohbfb7q_single_wht.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb9q_single_wht.fits
Writing out image to disk: ibohbfb9q_single_wht.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbgq_single_wht.fits
Writing out image to disk: ibohbfbgq_single_wht.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbdq_single_wht.fits
Writing out image to disk: ibohbfbdq_single_wht.fits
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.571706 to ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 2.011077 to ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = turbo
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbkq_single_sci.fits
-Generating simple FITS output: ibohbfbkq_single_sci.fits
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbpq_single_sci.fits
-Generating simple FITS output: ibohbfbpq_single_sci.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbkq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbpq_single_sci.fits
Writing out image to disk: ibohbfbkq_single_sci.fits
Writing out image to disk: ibohbfbpq_single_sci.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbkq_single_wht.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbpq_single_wht.fits
Writing out image to disk: ibohbfbkq_single_wht.fits
Writing out image to disk: ibohbfbpq_single_wht.fits
INFO:drizzlepac.util:==== Processing Step Separate Drizzle finished at 20:20:41.159 (02/12/2025)
==== Processing Step Separate Drizzle finished at 20:20:41.159 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Create Median started at 20:20:41.161 (02/12/2025)
==== Processing Step Create Median started at 20:20:41.161 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.createMedian:USER INPUT PARAMETERS for Create Median Step:
INFO:drizzlepac.createMedian: combine_bufsize : None
INFO:drizzlepac.createMedian: combine_grow : 1
INFO:drizzlepac.createMedian: combine_hthresh : None
INFO:drizzlepac.createMedian: combine_lthresh : None
INFO:drizzlepac.createMedian: combine_maskpt : 0.3
INFO:drizzlepac.createMedian: combine_nhigh : 1
INFO:drizzlepac.createMedian: combine_nlow : 0
INFO:drizzlepac.createMedian: combine_nsigma : 4 3
INFO:drizzlepac.createMedian: combine_type : median
INFO:drizzlepac.createMedian: compress : False
INFO:drizzlepac.createMedian: median : True
INFO:drizzlepac.createMedian: median_newmasks : True
INFO:drizzlepac.createMedian: proc_unit : native
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfb7q_flt.fits' is 0.0
reference sky value for image 'ibohbfb7q_flt.fits' is 0.0
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfb9q_flt.fits' is 1652.6279360108372
reference sky value for image 'ibohbfb9q_flt.fits' is 1652.6279360108372
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfbdq_flt.fits' is 4017.840039040874
reference sky value for image 'ibohbfbdq_flt.fits' is 4017.840039040874
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfbgq_flt.fits' is 232.48562545928166
reference sky value for image 'ibohbfbgq_flt.fits' is 232.48562545928166
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfbkq_flt.fits' is 914.2904827551788
reference sky value for image 'ibohbfbkq_flt.fits' is 914.2904827551788
INFO:drizzlepac.createMedian:reference sky value for image 'ibohbfbpq_flt.fits' is 3216.1809652750385
reference sky value for image 'ibohbfbpq_flt.fits' is 3216.1809652750385
INFO:drizzlepac.createMedian:Saving output median image to: 'ibohbf040_med.fits'
Saving output median image to: 'ibohbf040_med.fits'
INFO:drizzlepac.util:==== Processing Step Create Median finished at 20:20:41.42 (02/12/2025)
==== Processing Step Create Median finished at 20:20:41.42 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Blot started at 20:20:41.42 (02/12/2025)
==== Processing Step Blot started at 20:20:41.42 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.ablot:USER INPUT PARAMETERS for Blot Step:
INFO:drizzlepac.ablot: blot_addsky : True
INFO:drizzlepac.ablot: blot_interp : poly5
INFO:drizzlepac.ablot: blot_sinscl : 1.0
INFO:drizzlepac.ablot: blot_skyval : 0.0
INFO:drizzlepac.ablot: coeffs : True
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfb7q_flt.fits[sci,1]
Blot: creating blotted image: ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.000000 to blotted image ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfb7q_sci1_blt.fits
-Generating simple FITS output: ibohbfb7q_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb7q_sci1_blt.fits
Writing out image to disk: ibohbfb7q_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfb9q_flt.fits[sci,1]
Blot: creating blotted image: ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 1.033388 to blotted image ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfb9q_sci1_blt.fits
-Generating simple FITS output: ibohbfb9q_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfb9q_sci1_blt.fits
Writing out image to disk: ibohbfb9q_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfbdq_flt.fits[sci,1]
Blot: creating blotted image: ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 2.512355 to blotted image ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbdq_sci1_blt.fits
-Generating simple FITS output: ibohbfbdq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbdq_sci1_blt.fits
Writing out image to disk: ibohbfbdq_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfbgq_flt.fits[sci,1]
Blot: creating blotted image: ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.145373 to blotted image ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbgq_sci1_blt.fits
-Generating simple FITS output: ibohbfbgq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbgq_sci1_blt.fits
Writing out image to disk: ibohbfbgq_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfbkq_flt.fits[sci,1]
Blot: creating blotted image: ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 0.571706 to blotted image ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbkq_sci1_blt.fits
-Generating simple FITS output: ibohbfbkq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbkq_sci1_blt.fits
Writing out image to disk: ibohbfbkq_sci1_blt.fits
INFO:drizzlepac.ablot: Blot: creating blotted image: ibohbfbpq_flt.fits[sci,1]
Blot: creating blotted image: ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.ablot:Using default C-based coordinate transformation...
Using default C-based coordinate transformation...
INFO:drizzlepac.ablot:Applying sky value of 2.011077 to blotted image ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.outputimage:-Generating simple FITS output: ibohbfbpq_sci1_blt.fits
-Generating simple FITS output: ibohbfbpq_sci1_blt.fits
INFO:drizzlepac.outputimage:Writing out image to disk: ibohbfbpq_sci1_blt.fits
Writing out image to disk: ibohbfbpq_sci1_blt.fits
INFO:drizzlepac.util:==== Processing Step Blot finished at 20:20:42.470 (02/12/2025)
==== Processing Step Blot finished at 20:20:42.470 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Driz_CR started at 20:20:42.472 (02/12/2025)
==== Processing Step Driz_CR started at 20:20:42.472 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.drizCR:USER INPUT PARAMETERS for Driz_CR Step:
INFO:drizzlepac.drizCR: crbit : 4096
INFO:drizzlepac.drizCR: driz_cr : True
INFO:drizzlepac.drizCR: driz_cr_corr : False
INFO:drizzlepac.drizCR: driz_cr_ctegrow : 0
INFO:drizzlepac.drizCR: driz_cr_grow : 1
INFO:drizzlepac.drizCR: driz_cr_scale : 3.0 2.4
INFO:drizzlepac.drizCR: driz_cr_snr : 5.0 4.0
INFO:drizzlepac.drizCR: inmemory : False
INFO:drizzlepac.drizCR:Executing 4 parallel workers
INFO:drizzlepac.drizCR:Creating output: ibohbfb7q_sci1_crmask.fits
Creating output: ibohbfb7q_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ibohbfb9q_sci1_crmask.fits
Creating output: ibohbfb9q_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ibohbfbdq_sci1_crmask.fits
Creating output: ibohbfbdq_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ibohbfbgq_sci1_crmask.fits
Creating output: ibohbfbgq_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ibohbfbkq_sci1_crmask.fits
Creating output: ibohbfbkq_sci1_crmask.fits
INFO:drizzlepac.drizCR:Creating output: ibohbfbpq_sci1_crmask.fits
Creating output: ibohbfbpq_sci1_crmask.fits
INFO:drizzlepac.util:==== Processing Step Driz_CR finished at 20:20:44.725 (02/12/2025)
==== Processing Step Driz_CR finished at 20:20:44.725 (02/12/2025)
INFO:drizzlepac.util:==== Processing Step Final Drizzle started at 20:20:44.74 (02/12/2025)
==== Processing Step Final Drizzle started at 20:20:44.74 (02/12/2025)
INFO:drizzlepac.util:
INFO:drizzlepac.adrizzle:Interpreted paramDict with single=False as:
{'build': True, 'stepsize': 10, 'coeffs': True, 'wcskey': '', 'wht_type': 'EXP', 'kernel': 'square', 'wt_scl': 'exptime', 'pixfrac': 1.0, 'fillval': 'INDEF', 'maskval': None, 'bits': 528, 'units': 'cps'}
INFO:drizzlepac.adrizzle:USER INPUT PARAMETERS for Final Drizzle Step:
INFO:drizzlepac.adrizzle: bits : 528
INFO:drizzlepac.adrizzle: build : True
INFO:drizzlepac.adrizzle: clean : True
INFO:drizzlepac.adrizzle: coeffs : True
INFO:drizzlepac.adrizzle: crbit : 4096
INFO:drizzlepac.adrizzle: fillval : INDEF
INFO:drizzlepac.adrizzle: kernel : square
INFO:drizzlepac.adrizzle: logfile : astrodrizzle.log
INFO:drizzlepac.adrizzle: maskval : None
INFO:drizzlepac.adrizzle: pixfrac : 1.0
INFO:drizzlepac.adrizzle: proc_unit : native
INFO:drizzlepac.adrizzle: rules_file : None
INFO:drizzlepac.adrizzle: stepsize : 10
INFO:drizzlepac.adrizzle: units : cps
INFO:drizzlepac.adrizzle: wcskey :
INFO:drizzlepac.adrizzle: wht_type : EXP
INFO:drizzlepac.adrizzle: wt_scl : exptime
INFO:drizzlepac.adrizzle: **Using sub-sampling value of 10 for kernel square
INFO:drizzlepac.adrizzle:Running Drizzle to create output frame with WCS of:
INFO:astropy.wcs.wcs:WCS Keywords
WCS Keywords
INFO:astropy.wcs.wcs:
INFO:astropy.wcs.wcs:Number of WCS axes: 2
Number of WCS axes: 2
INFO:astropy.wcs.wcs:CTYPE : 'RA---TAN' 'DEC--TAN'
CTYPE : 'RA---TAN' 'DEC--TAN'
INFO:astropy.wcs.wcs:CUNIT : 'deg' 'deg'
CUNIT : 'deg' 'deg'
INFO:astropy.wcs.wcs:CRVAL : 189.12062959697133 62.23566816080676
CRVAL : 189.12062959697133 62.23566816080676
INFO:astropy.wcs.wcs:CRPIX : 546.5 482.0
CRPIX : 546.5 482.0
INFO:astropy.wcs.wcs:CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
CD1_1 CD1_2 : 2.5190728972332667e-05 2.5190629187110013e-05
INFO:astropy.wcs.wcs:CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
CD2_1 CD2_2 : 2.5190629187110013e-05 -2.5190728972332667e-05
INFO:astropy.wcs.wcs:NAXIS : 1093 964
NAXIS : 1093 964
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.000000 to ibohbfb7q_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 1.033388 to ibohbfb9q_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 2.512355 to ibohbfbdq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.145373 to ibohbfbgq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 0.571706 to ibohbfbkq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.adrizzle:-Drizzle input: ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.adrizzle:Applying sky value of 2.011077 to ibohbfbpq_flt.fits[sci,1]
INFO:drizzlepac.imageObject:Applying EXPTIME weighting to DQ mask for chip 1
INFO:drizzlepac.adrizzle:Using WCSLIB-based coordinate transformation...
INFO:drizzlepac.adrizzle:stepsize = 10
INFO:drizzlepac.cdriz:-Drizzling using kernel = square
INFO:drizzlepac.outputimage:-Generating multi-extension output file: ibohbf040_drz.fits
-Generating multi-extension output file: ibohbf040_drz.fits
INFO:drizzlepac.outputimage:Writing out to disk: ibohbf040_drz.fits
Writing out to disk: ibohbf040_drz.fits
WARNING:py.warnings:/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/astropy/io/fits/card.py:1036: VerifyWarning: Card is too long, comment will be truncated.
warnings.warn(
INFO:drizzlepac.util:==== Processing Step Final Drizzle finished at 20:20:46.597 (02/12/2025)
==== Processing Step Final Drizzle finished at 20:20:46.597 (02/12/2025)
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:AstroDrizzle Version 3.10.0 is finished processing at 20:20:46.599 (02/12/2025).
AstroDrizzle Version 3.10.0 is finished processing at 20:20:46.599 (02/12/2025).
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.astrodrizzle:
INFO:drizzlepac.util:
INFO:drizzlepac.util: -------------------- --------------------
-------------------- --------------------
INFO:drizzlepac.util: Step Elapsed time
Step Elapsed time
INFO:drizzlepac.util: -------------------- --------------------
-------------------- --------------------
INFO:drizzlepac.util:
INFO:drizzlepac.util: Initialization 0.6854 sec.
Initialization 0.6854 sec.
INFO:drizzlepac.util: Static Mask 0.1428 sec.
Static Mask 0.1428 sec.
INFO:drizzlepac.util: Subtract Sky 2.2893 sec.
Subtract Sky 2.2893 sec.
INFO:drizzlepac.util: Separate Drizzle 2.0715 sec.
Separate Drizzle 2.0715 sec.
INFO:drizzlepac.util: Create Median 0.2615 sec.
Create Median 0.2615 sec.
INFO:drizzlepac.util: Blot 1.0465 sec.
Blot 1.0465 sec.
INFO:drizzlepac.util: Driz_CR 2.2538 sec.
Driz_CR 2.2538 sec.
INFO:drizzlepac.util: Final Drizzle 1.8570 sec.
Final Drizzle 1.8570 sec.
INFO:drizzlepac.util: ==================== ====================
==================== ====================
INFO:drizzlepac.util: Total 10.6078 sec.
Total 10.6078 sec.
INFO:drizzlepac.util:
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfb7q_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfb9q_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfbdq_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfbgq_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfbkq_flt.fits
INFO:drizzlepac.imageObject:Removing intermediate files for ibohbfbpq_flt.fits
INFO:drizzlepac.util:Trailer file written to: astrodrizzle.log
Trailer file written to: astrodrizzle.log
The quality of the reprocessed DRZ product is significantly improved and the histogram of the background is narrower. Cosmic-rays which were present in the three reprocessed FLTs are effectively cleaned from the combined image.
drz_origdata = fits.getdata('orig/ibohbf040_drz.fits', ext=1)
drz_newdata = fits.getdata('ibohbf040_drz.fits', ext=1)
fig = plt.figure(figsize=(15, 8))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
ax1.imshow(drz_origdata[520:720, 750:970], vmin=0.4, vmax=0.6, cmap='Greys_r', origin='lower')
ax2.imshow(drz_newdata[520:720, 750:970], vmin=0.4, vmax=0.6, cmap='Greys_r', origin='lower')
ax1.set_title('Original DRZ', fontsize=20)
ax2.set_title('Reprocessed DRZ', fontsize=20)
Text(0.5, 1.0, 'Reprocessed DRZ')

fig = plt.figure(figsize=(15, 3))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
n, bins, patches = ax1.hist(drz_origdata.flatten(), bins=200, range=(0.4, 0.52))
n, bins, patches = ax2.hist(drz_newdata.flatten(), bins=200, range=(0.4, 0.52))
ax1.set_title('Original DRZ', fontsize=15)
ax2.set_title('Reprocessed DRZ', fontsize=15)
Text(0.5, 1.0, 'Reprocessed DRZ')
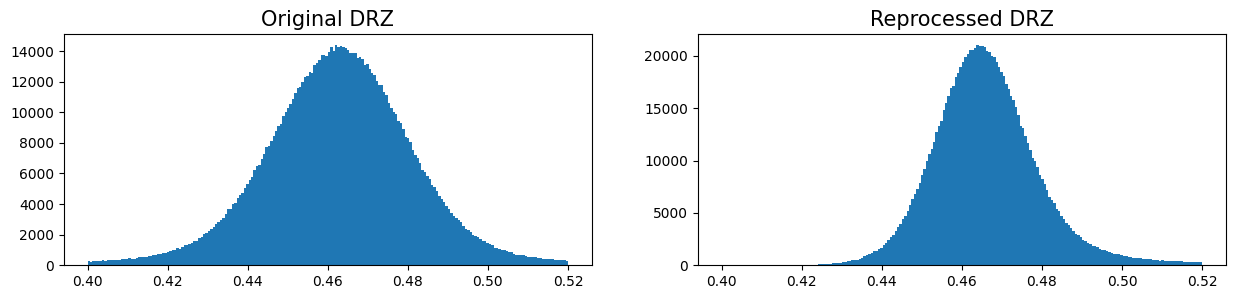
6. Conclusions#
Thank you for walking through this notebook. Now using WFC3 data, you should be more familiar with:
Analyzing exposure statistics for each read in an IMA file using
pstat.Reprocessing a single exposure and an image association using
calwf3.Combining the reprocessed exposures using
astrodrizzle.
Congratulations, you have completed the notebook!#
Additional Resources#
Below are some additional resources that may be helpful. Please send any questions through the HST Helpdesk.
-
see section 3.5.2 for reference to this notebook
see section 7.10 for further discussion of time-variable background
About this Notebook#
Authors: Jennifer Mack, Harish Khandrika; WFC3 Instrument Team
Created on: 2021-09-13
Updated on: 2023-11-16
Source: The notebook is sourced from hst_notebooks/notebooks/WFC3/calwf3_recalibration.
Citations#
If you use matplotlib, astropy, astroquery, drizzlepac, or wfc3tools for published research, please cite the
authors. Follow these links for more information about citing the libraries below:


