Sky Matching for HST Mosaics #
Learning Goals:#
In this tutorial we explore different options for handling the background sky with AstroDrizzle.
By the end of this notebook you will:
• Download data with astroquery
• Align data with TweakReg
• Compare background sky options using the AstroDrizzle parameter skymethod
Table of Contents#
1. Download the Observations with astroquery
1.1 Check image header data
1.2 Inspect the alignment
2. Align the visit-level drizzled data with TweakReg
2.1 Create a catalog of Gaia DR3 sources
2.2 Create a catalog of Gaia DR3 sources with Proper Motion Data
2.3 Run Tweakreg
2.4 Inspect the shift file and fit quality
2.5 Overplot matched sources and inspect fit residuals
2.6 Rerun TweakReg and update the header WCS
2.7 Run TweakBack to propogate the WCS to the FLT files
3. Compare skymethod options in AstroDrizzle
3.1 skymethod = 'localmin'
3.2 skymethod = 'match'
3.3 skymethod = 'globalmin+match'
3.4 skymethod = 'globalmin'
4. Compare the MDRIZSKY values for each method
5. Display the ‘sky matched’ science mosaic and weight image
6. Conclusion
Introduction #
When creating an image mosaic, AstroDrizzle has the ability to compute the sky and then either subtract or equalize the background in input images. Users may select the algorithm used for the sky subtraction via the skymethod parameter.
There are four methods available in sky matching: localmin, match, globalmin, and globalmin+match.
By applying drizzlepac.sky.sky(), or using the skymethod parameter in the call to drizzlepac.astrodrizzle.AstroDrizzle(), AstroDrizzle will update the keyword MDRIZSKY in the headers of the input files but it will not change the science data.
For images of sparse fields with few astronomical sources, the default skymethod = 'localmin' may be used, although this method can slightly oversubtract the background. For images with complicated backgrounds, such as nebulae and large host galaxies, skymethod = 'match' is recommended.
For more information on the specifics of this function, please refer to the documentation here
Below, each of the four methods is demonstrated using a single example dataset, and differences between the methods is highlighted.
# All imports needed through out this notebook are included at the beginning.
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
from collections import defaultdict
from IPython.display import clear_output
import glob
import os
import shutil
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import pandas
from astropy.coordinates import SkyCoord
from astropy.io import ascii, fits
from astropy.table import Table
from astropy.units import Quantity
import astropy.units as u
from astroquery.gaia import Gaia
from astroquery.mast import Observations
from drizzlepac import astrodrizzle, tweakback, tweakreg
from drizzlepac.haputils.astrometric_utils import create_astrometric_catalog
Gaia.MAIN_GAIA_TABLE = 'gaiadr3.gaia_source' # Change if different data release is desired
Gaia.ROW_LIMIT = 100000
1. Download the Observations with astroquery #
MAST queries may be done using query_criteria, where we specify:
–> obs_id, proposal_id, and filters
MAST data products may be downloaded by using download_products, where we specify:
–> products = calibrated (FLT, FLC) or drizzled (DRZ, DRC) files
–> type = standard products (CALxxx) or advanced products (HAP-SVM)
WFC3/IR observations of the Horsehead Nebula in the F160W filter obtained in HST proposal program 12812 will be used for this demonstration.
Nine visits were acquired in a 3x3 mosaic pattern on the sky, with two dither positions per visit in two IR filters. High level science products for these datasets were delivered to MAST in 2013, and this notebook is based on that user tutorial but has been updated to align these data to Gaia.
The 18 FLT images ibxl5*_flt.fits have been processed by the HST WFC3 pipeline (calwf3), which includes bias subtraction, dark current correction, cosmic-ray rejection, and flatfielding. The 9 DRZ files ibxl5*_drz.fits have been processed with AstroDrizzle to remove distortion and to combine the 2 dithered FLT frames by filter for each vist.
obs_ids = ['ibxl5*']
props = ['12812']
filts = ['F160W']
obsTable = Observations.query_criteria(obs_id=obs_ids, proposal_id=props, filters=filts)
products = Observations.get_product_list(obsTable)
data_prod = ['FLT', 'DRZ'] # ['FLC','FLT','DRC','DRZ']
data_type = ['CALWF3'] # ['CALACS','CALWF3','CALWP2','HAP-SVM']
Observations.download_products(products, download_dir='./science',
productSubGroupDescription=data_prod,
project=data_type)
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl50clq_flt.fits to ./science/mastDownload/HST/ibxl50clq/ibxl50clq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl50cqq_flt.fits to ./science/mastDownload/HST/ibxl50cqq/ibxl50cqq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl51eoq_flt.fits to ./science/mastDownload/HST/ibxl51eoq/ibxl51eoq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl51etq_flt.fits to ./science/mastDownload/HST/ibxl51etq/ibxl51etq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl52k0q_flt.fits to ./science/mastDownload/HST/ibxl52k0q/ibxl52k0q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl52k5q_flt.fits to ./science/mastDownload/HST/ibxl52k5q/ibxl52k5q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl53kxq_flt.fits to ./science/mastDownload/HST/ibxl53kxq/ibxl53kxq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl53l9q_flt.fits to ./science/mastDownload/HST/ibxl53l9q/ibxl53l9q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl54bgq_flt.fits to ./science/mastDownload/HST/ibxl54bgq/ibxl54bgq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl54blq_flt.fits to ./science/mastDownload/HST/ibxl54blq/ibxl54blq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl55f0q_flt.fits to ./science/mastDownload/HST/ibxl55f0q/ibxl55f0q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl55f5q_flt.fits to ./science/mastDownload/HST/ibxl55f5q/ibxl55f5q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl56huq_flt.fits to ./science/mastDownload/HST/ibxl56huq/ibxl56huq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl56i2q_flt.fits to ./science/mastDownload/HST/ibxl56i2q/ibxl56i2q_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl57adq_flt.fits to ./science/mastDownload/HST/ibxl57adq/ibxl57adq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl57aiq_flt.fits to ./science/mastDownload/HST/ibxl57aiq/ibxl57aiq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl58sqq_flt.fits to ./science/mastDownload/HST/ibxl58sqq/ibxl58sqq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl58svq_flt.fits to ./science/mastDownload/HST/ibxl58svq/ibxl58svq_flt.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl50030_drz.fits to ./science/mastDownload/HST/ibxl50030/ibxl50030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl51030_drz.fits to ./science/mastDownload/HST/ibxl51030/ibxl51030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl52030_drz.fits to ./science/mastDownload/HST/ibxl52030/ibxl52030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl53030_drz.fits to ./science/mastDownload/HST/ibxl53030/ibxl53030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl54030_drz.fits to ./science/mastDownload/HST/ibxl54030/ibxl54030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl55030_drz.fits to ./science/mastDownload/HST/ibxl55030/ibxl55030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl56030_drz.fits to ./science/mastDownload/HST/ibxl56030/ibxl56030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl57030_drz.fits to ./science/mastDownload/HST/ibxl57030/ibxl57030_drz.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibxl58030_drz.fits to ./science/mastDownload/HST/ibxl58030/ibxl58030_drz.fits ...
[Done]
| Local Path | Status | Message | URL |
|---|---|---|---|
| str55 | str8 | object | object |
| ./science/mastDownload/HST/ibxl50clq/ibxl50clq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl50cqq/ibxl50cqq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl51eoq/ibxl51eoq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl51etq/ibxl51etq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl52k0q/ibxl52k0q_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl52k5q/ibxl52k5q_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl53kxq/ibxl53kxq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl53l9q/ibxl53l9q_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl54bgq/ibxl54bgq_flt.fits | COMPLETE | None | None |
| ... | ... | ... | ... |
| ./science/mastDownload/HST/ibxl58svq/ibxl58svq_flt.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl50030/ibxl50030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl51030/ibxl51030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl52030/ibxl52030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl53030/ibxl53030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl54030/ibxl54030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl55030/ibxl55030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl56030/ibxl56030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl57030/ibxl57030_drz.fits | COMPLETE | None | None |
| ./science/mastDownload/HST/ibxl58030/ibxl58030_drz.fits | COMPLETE | None | None |
Move the files to the local working directory
files = glob.glob(os.path.join(os.curdir, 'science', 'mastDownload', 'HST', '*', '*fits'))
for im in files:
root = os.path.basename(im)
os.rename(im, './' + root)
if os.path.exists('./science'):
shutil.rmtree('science/')
1.1 Check image header data #
Here we will look at important keywords in the image headers.
files = sorted(glob.glob('*fl?.fits'))
keywords_ext0 = ["ROOTNAME", "ASN_ID", "TARGNAME", "DETECTOR", "FILTER", "EXPTIME",
"RA_TARG", "DEC_TARG", "POSTARG1", "POSTARG2", "DATE-OBS"]
keywords_ext1 = ["ORIENTAT"]
data = []
for file in files:
path_data = []
for keyword in keywords_ext0:
path_data.append(fits.getval(file, keyword, ext=0))
for keyword in keywords_ext1:
path_data.append(fits.getval(file, keyword, ext=1))
data.append(path_data)
keywords = keywords_ext0 + keywords_ext1
table = Table(np.array(data), names=keywords, dtype=['str', 'str', 'str', 'str', 'str', 'f8', 'f8', 'f8', 'f8', 'f8', 'str', 'f8'])
table['EXPTIME'].format = '7.1f'
table['RA_TARG'].format = table['DEC_TARG'].format = '7.4f'
table['POSTARG1'].format = table['POSTARG2'].format = '7.3f'
table['ORIENTAT'].format = '7.2f'
table.show_in_notebook()
1.2 Inspect the Alignment #
Check the active WCS solution in the image header. If the image is aligned to a catalog, list the number of matches and the fit RMS in mas.
Convert the fit RMS values to pixels for comparison with the alignment results performed later in this notebook.
ext_0_kws = ['DETECTOR']
ext_1_kws = ['WCSNAME', 'NMATCHES', 'RMS_RA', 'RMS_DEC']
det_scale = {'IR': 0.1283, 'UVIS': 0.0396, 'WFC': 0.05} # plate scale (arcsec/pixel)
format_dict = {}
col_dict = defaultdict(list)
for f in sorted(glob.glob('*dr?.fits')):
col_dict['FILENAME'].append(f)
hdr0 = fits.getheader(f, 0)
hdr1 = fits.getheader(f, 1)
for kw in ext_0_kws: # extension 0 keywords
col_dict[kw].append(hdr0[kw])
for kw in ext_1_kws: # extension 1 keywords
if 'RMS' in kw:
val = np.around(hdr1[kw], 1)
else:
val = hdr1[kw]
col_dict[kw].append(val)
for kw in ['RMS_RA', 'RMS_DEC']:
val = np.round(hdr1[kw]/1000./det_scale[hdr0['DETECTOR']], 2) # convert RMS from mas to pixels
col_dict[f'{kw}_pix'].append(val)
wcstable = Table(col_dict)
wcstable.show_in_notebook()
2. Align the visit-level drizzled data with TweakReg #
Here we will use TweakReg to align the DRZ files to Gaia DR3 and then use TweakBack to propagate those solutions back to the FLT image headers prior to combining with AstroDrizzle.
2.1 Create a catalog of Gaia DR3 sources #
This method uses the RA/Dec of the first image and a radius of 5’.
RA = table['RA_TARG'][0]
Dec = table['DEC_TARG'][0]
coord = SkyCoord(ra=RA, dec=Dec, unit=(u.deg, u.deg))
radius = Quantity(5., u.arcmin)
gaia_query = Gaia.query_object_async(coordinate=coord, radius=radius)
gaia_query
INFO: Query finished. [astroquery.utils.tap.core]
| solution_id | designation | source_id | random_index | ref_epoch | ra | ra_error | dec | dec_error | parallax | parallax_error | parallax_over_error | pm | pmra | pmra_error | pmdec | pmdec_error | ra_dec_corr | ra_parallax_corr | ra_pmra_corr | ra_pmdec_corr | dec_parallax_corr | dec_pmra_corr | dec_pmdec_corr | parallax_pmra_corr | parallax_pmdec_corr | pmra_pmdec_corr | astrometric_n_obs_al | astrometric_n_obs_ac | astrometric_n_good_obs_al | astrometric_n_bad_obs_al | astrometric_gof_al | astrometric_chi2_al | astrometric_excess_noise | astrometric_excess_noise_sig | astrometric_params_solved | astrometric_primary_flag | nu_eff_used_in_astrometry | pseudocolour | pseudocolour_error | ra_pseudocolour_corr | dec_pseudocolour_corr | parallax_pseudocolour_corr | pmra_pseudocolour_corr | pmdec_pseudocolour_corr | astrometric_matched_transits | visibility_periods_used | astrometric_sigma5d_max | matched_transits | new_matched_transits | matched_transits_removed | ipd_gof_harmonic_amplitude | ipd_gof_harmonic_phase | ipd_frac_multi_peak | ipd_frac_odd_win | ruwe | scan_direction_strength_k1 | scan_direction_strength_k2 | scan_direction_strength_k3 | scan_direction_strength_k4 | scan_direction_mean_k1 | scan_direction_mean_k2 | scan_direction_mean_k3 | scan_direction_mean_k4 | duplicated_source | phot_g_n_obs | phot_g_mean_flux | phot_g_mean_flux_error | phot_g_mean_flux_over_error | phot_g_mean_mag | phot_bp_n_obs | phot_bp_mean_flux | phot_bp_mean_flux_error | phot_bp_mean_flux_over_error | phot_bp_mean_mag | phot_rp_n_obs | phot_rp_mean_flux | phot_rp_mean_flux_error | phot_rp_mean_flux_over_error | phot_rp_mean_mag | phot_bp_rp_excess_factor | phot_bp_n_contaminated_transits | phot_bp_n_blended_transits | phot_rp_n_contaminated_transits | phot_rp_n_blended_transits | phot_proc_mode | bp_rp | bp_g | g_rp | radial_velocity | radial_velocity_error | rv_method_used | rv_nb_transits | rv_nb_deblended_transits | rv_visibility_periods_used | rv_expected_sig_to_noise | rv_renormalised_gof | rv_chisq_pvalue | rv_time_duration | rv_amplitude_robust | rv_template_teff | rv_template_logg | rv_template_fe_h | rv_atm_param_origin | vbroad | vbroad_error | vbroad_nb_transits | grvs_mag | grvs_mag_error | grvs_mag_nb_transits | rvs_spec_sig_to_noise | phot_variable_flag | l | b | ecl_lon | ecl_lat | in_qso_candidates | in_galaxy_candidates | non_single_star | has_xp_continuous | has_xp_sampled | has_rvs | has_epoch_photometry | has_epoch_rv | has_mcmc_gspphot | has_mcmc_msc | in_andromeda_survey | classprob_dsc_combmod_quasar | classprob_dsc_combmod_galaxy | classprob_dsc_combmod_star | teff_gspphot | teff_gspphot_lower | teff_gspphot_upper | logg_gspphot | logg_gspphot_lower | logg_gspphot_upper | mh_gspphot | mh_gspphot_lower | mh_gspphot_upper | distance_gspphot | distance_gspphot_lower | distance_gspphot_upper | azero_gspphot | azero_gspphot_lower | azero_gspphot_upper | ag_gspphot | ag_gspphot_lower | ag_gspphot_upper | ebpminrp_gspphot | ebpminrp_gspphot_lower | ebpminrp_gspphot_upper | libname_gspphot | dist |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| yr | deg | mas | deg | mas | mas | mas | mas / yr | mas / yr | mas / yr | mas / yr | mas / yr | mas | 1 / um | 1 / um | 1 / um | mas | deg | deg | deg | deg | deg | electron / s | electron / s | mag | electron / s | electron / s | mag | electron / s | electron / s | mag | mag | mag | mag | km / s | km / s | d | km / s | K | log(cm.s**-2) | dex | km / s | km / s | mag | mag | deg | deg | deg | deg | K | K | K | log(cm.s**-2) | log(cm.s**-2) | log(cm.s**-2) | dex | dex | dex | pc | pc | pc | mag | mag | mag | mag | mag | mag | mag | mag | mag | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| int64 | object | int64 | int64 | float64 | float64 | float32 | float64 | float32 | float64 | float32 | float32 | float32 | float64 | float32 | float64 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | int16 | bool | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | float32 | int16 | int16 | int16 | float32 | float32 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | bool | int16 | float64 | float32 | float32 | float32 | int16 | float64 | float32 | float32 | float32 | int16 | float64 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | int16 | int16 | int16 | int16 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | int16 | float32 | float32 | int16 | float32 | float32 | int16 | float32 | object | float64 | float64 | float64 | float64 | bool | bool | int16 | bool | bool | bool | bool | bool | bool | bool | bool | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | float32 | object | float64 |
| 1636148068921376768 | Gaia DR3 3216469053418858496 | 3216469053418858496 | 368024805 | 2016.0 | 85.25078206874738 | 0.34748474 | -2.444257621037541 | 0.31849745 | 0.9495336814954154 | 0.41641867 | 2.2802382 | 7.7956214 | -0.8302380599609497 | 0.43142426 | 7.751285021389771 | 0.36127582 | -0.072362915 | 0.0020754219 | -0.35186028 | 0.1278402 | -0.26970458 | 0.15042841 | -0.6164755 | 0.10007274 | 0.19517034 | -0.149899 | 357 | 0 | 357 | 0 | 1.8443948 | 436.133 | 1.3849133 | 2.208717 | 95 | False | -- | 1.233876 | 0.08958775 | -0.17215662 | -0.06320628 | -0.05079003 | -0.1254937 | 0.08907318 | 41 | 21 | 0.63760585 | 42 | 26 | 0 | 0.015478957 | 5.244358 | 0 | 0 | 1.0694424 | 0.20129156 | 0.24663325 | 0.2671826 | 0.5048746 | -80.303276 | -2.1296542 | -22.792238 | -43.76734 | False | 354 | 218.89926651301704 | 0.7818137 | 279.98904 | 19.836756 | 28 | 44.18486946908339 | 5.24969 | 8.416662 | 21.225359 | 36 | 308.8952727851156 | 11.356452 | 27.19998 | 18.523367 | 1.61298 | 0 | 2 | 0 | 3 | 0 | 2.701992 | 1.3886032 | 1.3133888 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.94547931488657 | -16.76434187915805 | 84.72852258013162 | -25.79667536635267 | False | False | 0 | False | False | False | False | False | False | False | False | 1.7621454e-13 | 5.111382e-13 | 0.99999774 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.009486679186673242 | |
| 1636148068921376768 | Gaia DR3 3216469057714526080 | 3216469057714526080 | 160562994 | 2016.0 | 85.24865858921503 | 0.0807162 | -2.441354584616136 | 0.07413533 | 0.24702429233455972 | 0.09551564 | 2.5862184 | 1.38756 | 0.7312738742365584 | 0.09950856 | 1.1792206068277042 | 0.08531875 | -0.09268238 | -0.016587587 | -0.35330772 | 0.15432145 | -0.25057435 | 0.17109188 | -0.649208 | 0.11624458 | 0.24781711 | -0.1273712 | 389 | 0 | 387 | 2 | -1.0886481 | 412.40457 | 0.0 | 0.0 | 95 | False | -- | 1.2642988 | 0.020441843 | -0.14009616 | -0.07836674 | 0.0031425122 | -0.10781701 | 0.09727073 | 45 | 21 | 0.1476955 | 46 | 29 | 0 | 0.021776078 | 2.3582678 | 0 | 0 | 0.9599596 | 0.19218576 | 0.2656598 | 0.22300626 | 0.48614076 | -98.501114 | -2.7790413 | -25.351597 | -43.091904 | False | 385 | 1533.7964710362219 | 1.6690606 | 918.95795 | 17.722948 | 45 | 414.8798543565718 | 7.5229797 | 55.148342 | 18.793736 | 44 | 1747.7167079736257 | 11.556245 | 151.23569 | 16.641718 | 1.4099631 | 0 | 1 | 0 | 1 | 0 | 2.1520176 | 1.0707874 | 1.0812302 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.94176557869633 | -16.764874991852714 | 84.72628577778967 | -25.793696653062412 | False | False | 0 | False | False | False | False | False | True | True | False | 1.2362927e-13 | 6.260371e-13 | 0.9999927 | 6097.917 | 5930.6045 | 6142.094 | 4.5226 | 4.5009 | 4.5768 | -3.9361 | -4.0965 | -3.6061 | 919.4193 | 833.37 | 948.0848 | 3.6223 | 3.5315 | 3.6515 | 2.7407 | 2.6619 | 2.7644 | 1.5455 | 1.5014 | 1.5587 | MARCS | 0.013011650487557274 |
| 1636148068921376768 | Gaia DR3 3216467855123686016 | 3216467855123686016 | 668710222 | 2016.0 | 85.25474417063919 | 0.07255654 | -2.470084090608095 | 0.06763807 | 1.705994308777196 | 0.09274939 | 18.39359 | 13.672125 | -3.181681654139363 | 0.08841129 | -13.296762516546895 | 0.07709159 | -0.12809613 | 0.08502215 | -0.3073343 | 0.19231312 | -0.37712017 | 0.17119895 | -0.66374385 | 0.03020479 | 0.40648898 | -0.117953785 | 376 | 0 | 376 | 0 | 4.4143248 | 528.08984 | 0.39314708 | 4.8827662 | 31 | False | 1.3003229 | -- | -- | -- | -- | -- | -- | -- | 43 | 21 | 0.13255168 | 45 | 26 | 0 | 0.027088897 | 14.045337 | 0 | 2 | 1.1654105 | 0.22388098 | 0.3077812 | 0.25345388 | 0.42603984 | -72.65342 | -1.7500962 | -25.84865 | -41.594986 | False | 368 | 2476.027650243042 | 4.3828444 | 564.9362 | 17.202978 | 40 | 585.7780381608451 | 11.731945 | 49.930172 | 18.41921 | 36 | 2981.23630414607 | 24.774414 | 120.33529 | 16.061905 | 1.4406197 | 0 | 4 | 0 | 3 | 0 | 2.3573055 | 1.2162323 | 1.1410732 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.9712942044114 | -16.772811629675818 | 84.73186766212942 | -25.822629314586116 | False | False | 0 | True | False | False | False | False | False | True | False | 1.1540595e-13 | 1.3798553e-09 | 0.9999877 | 5968.2295 | 5923.5728 | 5995.96 | 4.6275 | 4.589 | 4.6636 | -4.0538 | -4.1164 | -3.7235 | 542.7187 | 531.214 | 555.837 | 4.0502 | 4.0231 | 4.069 | 3.0096 | 2.9875 | 3.0238 | 1.7022 | 1.6897 | 1.7104 | MARCS | 0.017083879996516314 |
| 1636148068921376768 | Gaia DR3 3216469019060675328 | 3216469019060675328 | 755626735 | 2016.0 | 85.26833524857398 | 0.45910412 | -2.4416029853336667 | 0.40957135 | -1.0905664099598757 | 0.530555 | -2.0555198 | 1.1219171 | 0.8643062489270098 | 0.5828771 | 0.715313093573643 | 0.47160438 | -0.054446124 | -0.015339496 | -0.33468443 | 0.13777287 | -0.2609537 | 0.1763433 | -0.5611049 | 0.031097142 | 0.2177652 | -0.11443702 | 330 | 0 | 329 | 1 | 1.0299137 | 382.76816 | 0.68933684 | 0.29942906 | 95 | False | -- | 1.2502365 | 0.123004876 | -0.19324453 | -0.12574936 | 0.032932058 | -0.14361922 | 0.064552374 | 39 | 19 | 0.84676933 | 41 | 25 | 0 | 0.06055019 | 12.78158 | 0 | 0 | 1.0397482 | 0.20912738 | 0.2604514 | 0.24491833 | 0.5289087 | -85.225365 | -3.2899575 | -29.32366 | -42.54999 | False | 328 | 147.74586838835364 | 0.72871625 | 202.74814 | 20.263578 | 29 | 36.02716968454987 | 3.8942373 | 9.251406 | 21.446966 | 37 | 190.87637950900836 | 7.5462556 | 25.294184 | 19.046015 | 1.5357692 | 0 | 0 | 0 | 0 | 0 | 2.4009514 | 1.1833878 | 1.2175636 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.9515174309682 | -16.74757367009055 | 84.74809544110592 | -25.794662806626693 | False | False | 0 | False | False | False | False | False | False | False | False | 7.747473e-13 | 5.229698e-13 | 0.99999446 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.017945489552972947 | |
| 1636148068921376768 | Gaia DR3 3216468611037930624 | 3216468611037930624 | 935473409 | 2016.0 | 85.2386085747217 | 0.46767473 | -2.462292300986681 | 0.4031481 | 0.7770076972461502 | 0.59839076 | 1.2984954 | 5.413292 | 0.5531465058042339 | 0.6129275 | -5.384956673257242 | 0.49430868 | 0.014938566 | 0.06448122 | -0.21245089 | 0.0711942 | -0.2614386 | 0.1208418 | -0.5615382 | -0.026767768 | 0.38506997 | -0.09344254 | 342 | 0 | 342 | 0 | 0.9642493 | 406.29196 | 1.4710811 | 1.2780555 | 95 | False | -- | 1.1623133 | 0.12403801 | -0.19795567 | -0.079425685 | 0.06298836 | -0.106788404 | 0.1012403 | 39 | 17 | 0.8698572 | 40 | 24 | 0 | 0.036293596 | 90.33434 | 0 | 0 | 1.0364223 | 0.1608992 | 0.30992013 | 0.18475303 | 0.44911218 | -82.79225 | -4.6845818 | -28.746756 | -41.727596 | False | 341 | 137.96486675397304 | 0.78542095 | 175.65723 | 20.337946 | 21 | 59.02707720646006 | 6.8534393 | 8.612767 | 20.910913 | 33 | 211.43949067561897 | 9.650035 | 21.91075 | 18.93493 | 1.9604018 | 0 | 1 | 0 | 1 | 0 | 1.9759827 | 0.5729675 | 1.4030151 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.95627609646655 | -16.78348111875944 | 84.71428834509156 | -25.814252456150466 | False | False | 0 | False | False | False | False | False | False | False | False | 1.2971835e-13 | 8.269208e-13 | 0.9999991 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.01836293659726027 | |
| 1636148068921376768 | Gaia DR3 3216468675465488256 | 3216468675465488256 | 16647219 | 2016.0 | 85.23467309871081 | 0.51438564 | -2.4498085573057633 | 0.43504587 | 0.33731643747067025 | 0.63177866 | 0.5339156 | 2.3331635 | 2.3330716928916786 | 0.65796125 | 0.02069617827588948 | 0.5042113 | -0.011451531 | -0.19442908 | -0.41642103 | 0.03437781 | -0.38334745 | 0.10441229 | -0.6943009 | 0.1926607 | 0.4085249 | -0.12117048 | 334 | 0 | 333 | 1 | 0.65179557 | 368.28964 | 1.3423299 | 1.2644218 | 95 | False | -- | 1.3347516 | 0.11629938 | -0.2020829 | -0.07508502 | -0.014079772 | -0.13441803 | 0.09302474 | 38 | 17 | 0.95642906 | 39 | 25 | 0 | 0.0040856865 | 64.82368 | 0 | 0 | 1.0245674 | 0.20001507 | 0.36711606 | 0.26003 | 0.40765452 | -98.68751 | 4.1797576 | -20.715187 | -39.459377 | False | 327 | 151.8625314562344 | 0.66563815 | 228.14578 | 20.23374 | 21 | 36.17745844357809 | 5.4620037 | 6.6234775 | 21.442448 | 29 | 204.66180430023923 | 8.290443 | 24.686472 | 18.970303 | 1.5859032 | 0 | 0 | 0 | 0 | 0 | 2.472145 | 1.2087078 | 1.2634373 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.94281943924076 | -16.781174971492643 | 84.71043259626695 | -25.801632853854667 | False | False | 0 | False | False | False | False | False | False | False | False | 4.767196e-13 | 5.8040146e-11 | 0.99999815 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.020028009828231417 | |
| 1636148068921376768 | Gaia DR3 3216469023354785408 | 3216469023354785408 | 176691911 | 2016.0 | 85.27075582329469 | 0.1961405 | -2.441307234608435 | 0.17860357 | 0.6636681928453716 | 0.2341562 | 2.8342967 | 6.585025 | 6.575404865804311 | 0.24365711 | -0.35580991638604564 | 0.20158039 | -0.04231842 | 0.014788889 | -0.3154728 | 0.11617606 | -0.2603726 | 0.12642686 | -0.5700941 | 0.13072464 | 0.23919845 | -0.061890464 | 368 | 0 | 366 | 2 | 0.43652844 | 405.4187 | 0.072520114 | 0.015899755 | 95 | False | -- | 1.2586821 | 0.05131671 | -0.1420974 | -0.08405918 | -0.029754888 | -0.114538334 | 0.13519505 | 43 | 20 | 0.3494921 | 45 | 28 | 0 | 0.018798698 | 6.73902 | 0 | 0 | 1.0153817 | 0.21722564 | 0.24762863 | 0.26764935 | 0.5151869 | -74.446075 | -3.9929616 | -22.67545 | -43.95803 | False | 363 | 409.71337850914114 | 1.0203439 | 401.5444 | 19.156166 | 37 | 65.82193793586049 | 5.4644194 | 12.04555 | 20.792616 | 40 | 530.0466563553375 | 7.329029 | 72.32154 | 17.93711 | 1.4543548 | 0 | 1 | 0 | 1 | 0 | 2.855505 | 1.6364498 | 1.2190552 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.95241508079897 | -16.745293946283045 | 84.75079163706141 | -25.7944553638111 | False | False | 0 | False | False | False | False | False | False | False | False | 3.5761834e-13 | 5.110677e-13 | 0.999994 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.020043830041751424 | |
| 1636148068921376768 | Gaia DR3 3216467855122414464 | 3216467855122414464 | 923936172 | 2016.0 | 85.25578005166672 | 0.50923383 | -2.4765054474694934 | 0.45520246 | 0.7769168571521851 | 0.5960472 | 1.3034484 | 0.9249258 | 0.8018697645542915 | 0.6797768 | 0.4609692420907058 | 0.5313265 | -0.11263442 | 0.11976503 | -0.22806385 | 0.19673494 | -0.18776836 | 0.22860377 | -0.61896616 | 0.19363396 | 0.23427469 | -0.13258351 | 285 | 0 | 282 | 3 | -0.40685338 | 300.89392 | 0.0 | 2.4194166e-15 | 95 | False | -- | 1.0979824 | 0.14102378 | -0.12671481 | -0.15378633 | -0.059823737 | -0.18096454 | 0.24567267 | 33 | 18 | 0.9812631 | 34 | 19 | 0 | 0.07628962 | 4.733895 | 0 | 0 | 0.98153275 | 0.21344621 | 0.29472753 | 0.19238599 | 0.43329495 | -87.93401 | -1.0898006 | -29.688093 | -42.996693 | False | 280 | 136.19056298917084 | 0.83884525 | 162.35481 | 20.352 | 23 | 45.240498301565275 | 8.204231 | 5.5142884 | 21.199724 | 31 | 151.9858001141609 | 8.52438 | 17.829544 | 19.293388 | 1.4481642 | 0 | 0 | 0 | 2 | 0 | 1.9063358 | 0.8477249 | 1.0586109 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.97773758997462 | -16.774872090351703 | 84.73275587575476 | -25.829084209901822 | False | False | 0 | False | False | False | False | False | False | False | False | 1.0344582e-09 | 1.8264745e-10 | 0.99999684 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.023539736369753695 | |
| 1636148068921376768 | Gaia DR3 3216468164360052992 | 3216468164360052992 | 23546710 | 2016.0 | 85.27451682697576 | 2.2556965 | -2.472305714228985 | 1.5849423 | -- | -- | -- | -- | -- | -- | -- | -- | 0.536678 | -- | -- | -- | -- | -- | -- | -- | -- | -- | 80 | 0 | 80 | 0 | 1.4301157 | 93.14176 | 0.0 | 0.0 | 3 | False | -- | -- | -- | -- | -- | -- | -- | -- | 10 | 7 | 4.1676683 | 10 | 6 | 0 | 0.09136518 | 171.88559 | 0 | 0 | -- | 0.3697472 | 0.50056857 | 0.41110742 | 0.52664024 | -76.56397 | -22.63934 | -33.053043 | -39.32441 | False | 79 | 78.44416585717141 | 1.3150449 | 59.651325 | 20.950966 | 4 | 66.57637510952588 | 25.076605 | 2.6549199 | 20.780241 | 5 | 135.564179093732 | 25.503525 | 5.3155074 | 19.417534 | 2.5768716 | 0 | 0 | 0 | 0 | 0 | 1.3627071 | -0.17072487 | 1.533432 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.98291592741373 | -16.75633924939091 | 84.75370889485112 | -25.82557011527804 | False | False | 0 | False | False | False | False | False | False | False | False | 2.4502437e-05 | 1.4911667e-05 | 0.99836755 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.02782140743048771 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1636148068921376768 | Gaia DR3 3216456065439222912 | 3216456065439222912 | 1421695677 | 2016.0 | 85.31116982575814 | 0.40173137 | -2.5106681948456453 | 0.4013123 | 1.736133118826882 | 0.45418435 | 3.8225296 | 9.741213 | -5.152917907458617 | 0.517365 | -8.266719879098039 | 0.443483 | -0.16442835 | -0.076135896 | -0.43010193 | 0.24256699 | 0.0072904215 | 0.35669935 | -0.7236328 | 0.12072589 | -0.0053872755 | -0.32701382 | 290 | 0 | 289 | 1 | 3.1741064 | 417.32562 | 1.6136845 | 3.0210483 | 95 | False | -- | 1.1419736 | 0.1037138 | -0.102503575 | -0.23006625 | -0.033000063 | -0.16398549 | 0.22349931 | 34 | 20 | 0.8391437 | 37 | 25 | 0 | 0.02747992 | 177.14145 | 1 | 0 | 1.1351129 | 0.1884894 | 0.27369884 | 0.21929786 | 0.48222566 | -80.36146 | -4.405813 | -22.027073 | -42.93724 | False | 287 | 234.3866961283912 | 0.9647052 | 242.96199 | 19.762535 | 24 | 75.9456305925942 | 9.633069 | 7.883846 | 20.637285 | 29 | 334.7660203080705 | 11.112565 | 30.125 | 18.436043 | 1.7522823 | 0 | 2 | 0 | 1 | 0 | 2.2012424 | 0.87475014 | 1.3264923 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 207.0361400496104 | -16.74167521407987 | 84.79282428995414 | -25.865235525984968 | False | False | 0 | False | False | False | False | False | False | False | False | 6.410813e-12 | 2.0735247e-09 | 0.9999794 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.080839193349907 | |
| 1636148068921376768 | Gaia DR3 3216472558112887808 | 3216472558112887808 | 796706940 | 2016.0 | 85.18852621317357 | 0.2259397 | -2.405656267543347 | 0.21768318 | -0.11388777731196244 | 0.28216147 | -0.40362626 | 1.2198783 | 1.184294234555146 | 0.26978385 | -0.29248995206570705 | 0.2340413 | -0.26953048 | 0.1911443 | -0.23510878 | 0.32237884 | -0.36343005 | 0.30793124 | -0.65272945 | 0.037301254 | 0.33524925 | -0.26868442 | 325 | 0 | 325 | 0 | -0.717088 | 325.74405 | 0.0 | 0.0 | 95 | False | -- | 1.2839941 | 0.056570467 | -0.19961677 | 0.015470109 | -0.18636444 | -0.09788927 | 0.019295616 | 38 | 20 | 0.43101293 | 38 | 23 | 0 | 0.030880287 | 8.535898 | 0 | 0 | 0.97071016 | 0.22002906 | 0.28557843 | 0.20794256 | 0.4101255 | -65.66478 | 0.8588567 | -24.406776 | -43.698612 | False | 320 | 364.00507922068095 | 0.90043736 | 404.25363 | 19.284597 | 34 | 137.15507118818775 | 11.79715 | 11.62612 | 19.995512 | 31 | 342.4081435440125 | 7.838386 | 43.683502 | 18.411535 | 1.3174629 | 0 | 1 | 0 | 1 | 0 | 1.5839767 | 0.7109146 | 0.87306213 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.87962201372886 | -16.801535222612248 | 84.66107447497244 | -25.75581047640869 | False | False | 0 | False | False | False | False | False | False | False | False | 2.1622872e-10 | 1.3842859e-10 | 0.99999607 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08112646296289025 | |
| 1636148068921376768 | Gaia DR3 3216444937178204160 | 3216444937178204160 | 1386239099 | 2016.0 | 85.19030586577739 | 0.07698814 | -2.5028819323642013 | 0.06952155 | 0.49047638721509285 | 0.09691278 | 5.0610085 | 9.442557 | 7.014016212995759 | 0.09374031 | -6.321825218787011 | 0.079626285 | -0.054927044 | 0.02979098 | -0.2849793 | 0.09145997 | -0.34964725 | 0.09440442 | -0.62761176 | 0.066617794 | 0.35496876 | -0.08418727 | 381 | 0 | 381 | 0 | 2.634945 | 476.3032 | 0.30653253 | 2.2057276 | 95 | False | -- | 1.4101175 | 0.020281127 | -0.15829301 | -0.032261044 | -0.025952134 | -0.049249224 | 0.06220646 | 43 | 21 | 0.13505712 | 43 | 25 | 0 | 0.023879923 | 9.0169325 | 1 | 0 | 1.0968245 | 0.20904742 | 0.29357752 | 0.24537306 | 0.4592325 | -93.210556 | 0.81500864 | -25.391544 | -42.085728 | False | 373 | 1818.6555646912966 | 1.5091 | 1205.126 | 17.53799 | 38 | 663.8714987983407 | 11.664461 | 56.914032 | 18.283333 | 38 | 1679.6880569559742 | 12.2571 | 137.03796 | 16.684824 | 1.288622 | 0 | 2 | 0 | 0 | 0 | 1.5985088 | 0.74534225 | 0.8531666 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.97046839266432 | -16.845058105618648 | 84.65904656064926 | -25.85303525756708 | False | False | 0 | True | False | False | False | False | True | True | False | 2.1057866e-11 | 5.2152347e-13 | 0.9999929 | 4720.8716 | 4589.9756 | 4839.4907 | 4.6567 | 4.5776 | 4.7072 | -2.2392 | -2.4267 | -1.7745 | 862.0521 | 812.244 | 948.8539 | 1.0476 | 0.9054 | 1.2078 | 0.7958 | 0.685 | 0.9228 | 0.4362 | 0.3746 | 0.5038 | PHOENIX | 0.08121814053391099 |
| 1636148068921376768 | Gaia DR3 3216468782835372032 | 3216468782835372032 | 657511649 | 2016.0 | 85.17292618358479 | 0.19054802 | -2.4623957493034743 | 0.17272547 | 2.1624020293821835 | 0.23938343 | 9.0332155 | 2.4493213 | -2.037035196031785 | 0.2268017 | 1.3600228792903124 | 0.19178832 | -0.14530747 | 0.112551525 | -0.20920323 | 0.17061059 | -0.3053354 | 0.17149667 | -0.5708101 | 0.043384105 | 0.30095217 | -0.20159128 | 352 | 0 | 351 | 1 | -0.48073354 | 416.80234 | 0.59515935 | 1.1965727 | 95 | False | -- | 1.0901086 | 0.05042796 | -0.17146336 | -0.0065654744 | -0.110863455 | -0.030477438 | 0.0075620045 | 40 | 20 | 0.33940324 | 40 | 23 | 0 | 0.030575372 | 178.93437 | 0 | 0 | 0.9807946 | 0.16747355 | 0.28867003 | 0.17314363 | 0.43588594 | -74.866936 | 5.0166435 | -23.469093 | -43.28577 | False | 347 | 435.6715921697949 | 1.0631166 | 409.80606 | 19.089468 | 26 | 44.39955876616971 | 6.0446095 | 7.3453145 | 21.220095 | 38 | 673.5576808594151 | 8.575238 | 78.54682 | 17.676958 | 1.6479322 | 0 | 0 | 0 | 0 | 0 | 3.5431366 | 2.1306267 | 1.4125099 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.92457804544165 | -16.841666317801355 | 84.64143853749016 | -25.811932421839387 | False | False | 0 | False | False | False | False | False | False | False | False | 2.7495896e-13 | 5.0978774e-13 | 0.99999624 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08200148643415694 | |
| 1636148068921376768 | Gaia DR3 3216455691775435392 | 3216455691775435392 | 1079684357 | 2016.0 | 85.30495708910514 | 1.4646537 | -2.5178904392586623 | 1.198452 | 1.0621429300659084 | 1.496934 | 0.70954555 | 14.06971 | 2.362959871277592 | 1.4997277 | -13.869864598271748 | 1.2972544 | 0.41949913 | 0.5497209 | -0.122153305 | -0.007439146 | 0.36512154 | 0.13959806 | -0.49781814 | 0.08308936 | -0.12201575 | 0.1388765 | 113 | 0 | 112 | 1 | -1.8722427 | 109.50301 | 0.0 | 0.0 | 95 | False | -- | 1.1160979 | 0.38008702 | 0.2803165 | 0.108602926 | 0.3289834 | 0.09997459 | 0.17758605 | 13 | 11 | 2.1380084 | 14 | 8 | 0 | 0.13895129 | 172.40553 | 0 | 0 | 0.87120515 | 0.36661693 | 0.3774008 | 0.22178955 | 0.376025 | -25.664597 | -18.63486 | -15.046993 | 42.01079 | False | 111 | 83.73466570505262 | 0.9941574 | 84.22677 | 20.880104 | 8 | 48.316372981047714 | 11.551536 | 4.1826797 | 21.128307 | 11 | 143.12426368829165 | 13.869087 | 10.31966 | 19.358612 | 2.286277 | 0 | 0 | 0 | 0 | 1 | 1.7696953 | 0.24820328 | 1.521492 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 207.03981962770035 | -16.750522211440767 | 84.7856405966112 | -25.872228605607752 | False | False | 0 | False | False | False | False | False | False | False | False | 1.2387219e-10 | 4.4580528e-08 | 0.99999696 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08219069174617054 | |
| 1636148068921376768 | Gaia DR3 3216469847988496640 | 3216469847988496640 | 1750321072 | 2016.0 | 85.33205874915119 | 0.4250795 | -2.425249860759321 | 0.37327173 | 0.22421272188486763 | 0.48120424 | 0.46594086 | 1.1375393 | 0.6146611315465014 | 0.53041315 | -0.9571766658995218 | 0.43026003 | -0.08336612 | -0.04359924 | -0.3503951 | 0.17059404 | -0.22215058 | 0.20433807 | -0.63215035 | 0.105049506 | 0.21047379 | -0.11205828 | 395 | 0 | 393 | 2 | 3.2212033 | 549.23535 | 1.7964642 | 2.885576 | 95 | False | -- | 1.0470759 | 0.107746 | -0.16536176 | -0.1214191 | 0.04378124 | -0.12220128 | 0.12459754 | 46 | 21 | 0.7767407 | 50 | 31 | 0 | 0.016963525 | 142.54533 | 0 | 0 | 1.1170963 | 0.21057051 | 0.30917907 | 0.26333737 | 0.43816993 | -90.41127 | -5.9151006 | -22.575792 | -42.194275 | False | 384 | 176.802211701999 | 0.77002114 | 229.60695 | 20.068647 | 21 | 48.094409264223955 | 8.200751 | 5.8646345 | 21.133307 | 41 | 300.8747652262563 | 8.945304 | 33.63494 | 18.551931 | 1.9737829 | 0 | 1 | 0 | 2 | 0 | 2.5813751 | 1.0646591 | 1.516716 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.96721911695258 | -16.683584284834694 | 84.81941412521121 | -25.78062488637266 | False | False | 0 | False | False | False | False | False | False | False | False | 5.8075224e-13 | 6.0352385e-13 | 0.99999946 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08234330900782977 | |
| 1636148068921376768 | Gaia DR3 3216470840133093504 | 3216470840133093504 | 400465001 | 2016.0 | 85.30747238821475 | 0.29184374 | -2.38949359231377 | 0.26074883 | 0.12019260367543594 | 0.33985 | 0.35366368 | 5.1894574 | 1.7876459824526552 | 0.36385092 | -4.871836205810002 | 0.2897296 | -0.09265526 | -0.0014098006 | -0.31270424 | 0.17439121 | -0.19470231 | 0.19349204 | -0.57027817 | 0.14670835 | 0.18696235 | -0.098352544 | 363 | 0 | 360 | 3 | 0.86954176 | 431.9599 | 0.8580658 | 1.1291534 | 95 | False | -- | 1.1637046 | 0.07667623 | -0.1388337 | -0.07941982 | -0.006895077 | -0.116873085 | 0.1331642 | 43 | 20 | 0.52750075 | 45 | 28 | 0 | 0.0151079735 | 6.0423245 | 0 | 0 | 1.031905 | 0.20595434 | 0.25317675 | 0.2603947 | 0.5014082 | -76.88399 | -3.9765499 | -21.605133 | -43.848877 | False | 357 | 265.88909242259996 | 0.897993 | 296.0926 | 19.625616 | 34 | 51.091312482354034 | 6.8466034 | 7.462286 | 21.067675 | 41 | 406.2893467231781 | 8.791597 | 46.21337 | 18.225807 | 1.7201934 | 0 | 0 | 0 | 2 | 0 | 2.8418674 | 1.4420586 | 1.3998089 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 206.92225446615564 | -16.6887610547889 | 84.7935869203766 | -25.744006573347082 | False | False | 0 | False | False | False | False | False | False | False | False | 8.483602e-13 | 5.134769e-13 | 0.9999985 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08269446496314736 | |
| 1636148068921376768 | Gaia DR3 3216443872025058816 | 3216443872025058816 | 925732867 | 2016.0 | 85.24559042309376 | 0.71123976 | -2.535389226550673 | 0.73737574 | 0.8980238349417092 | 0.9670169 | 0.9286537 | 1.5929452 | -1.4791026414262343 | 0.8477286 | -0.5913795705051671 | 0.7786666 | -0.2839063 | 0.3210551 | -0.23774688 | 0.2605932 | -0.51915574 | 0.22001855 | -0.61014396 | -0.19906428 | 0.4816959 | -0.21136881 | 217 | 0 | 215 | 2 | 2.8487191 | 283.3687 | 1.5834634 | 1.1608537 | 95 | False | -- | 1.6825651 | 0.205878 | -0.3583018 | 0.26714784 | -0.16991253 | -0.030751199 | -0.10089172 | 25 | 16 | 1.3951224 | 26 | 13 | 0 | 0.004611447 | 65.27896 | 0 | 0 | 1.1408558 | 0.35099515 | 0.18764296 | 0.354644 | 0.6033324 | -63.270237 | 8.353648 | -27.99232 | -44.789764 | False | 213 | 116.43417552584557 | 0.8913834 | 130.62187 | 20.522165 | 16 | 50.024253993893815 | 9.838917 | 5.0843253 | 21.090591 | 22 | 124.74988720000201 | 8.939355 | 13.955133 | 19.507795 | 1.5010554 | 0 | 0 | 0 | 0 | 0 | 1.5827961 | 0.56842613 | 1.01437 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 207.02730810483163 | -16.811187434288154 | 84.71905604380503 | -25.88755596761321 | False | False | 0 | False | False | False | False | False | False | False | False | 1.085153e-07 | 5.787982e-09 | 0.9999933 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08286217986852269 | |
| 1636148068921376768 | Gaia DR3 3216455653128695936 | 3216455653128695936 | 1467422921 | 2016.0 | 85.29328038861492 | 0.2429748 | -2.526244532656098 | 0.23323357 | 7.187770844565762 | 0.2993434 | 24.01179 | 36.290695 | -33.9237172532513 | 0.2799555 | 12.891699735107602 | 0.25018305 | -0.0739246 | 0.051945604 | -0.34340528 | 0.11745921 | -0.23482792 | 0.14009184 | -0.67943627 | -0.004798767 | 0.24896738 | -0.115868084 | 302 | 0 | 300 | 2 | 0.91878456 | 374.8465 | 0.9050687 | 2.162002 | 95 | False | -- | 1.15316 | 0.06230389 | -0.1844487 | -0.073425606 | -0.014452084 | -0.012071895 | 0.048439857 | 35 | 20 | 0.4237814 | 38 | 26 | 0 | 0.018915895 | 11.662993 | 0 | 0 | 1.0369805 | 0.22564329 | 0.2900915 | 0.27226767 | 0.43290338 | -72.82827 | 0.47325253 | -21.18871 | -44.038074 | False | 291 | 415.48303554301236 | 1.1920303 | 348.55072 | 19.140984 | 26 | 54.11411024598962 | 8.189754 | 6.607538 | 21.005266 | 35 | 627.9098762932185 | 13.446248 | 46.697777 | 17.753153 | 1.6415206 | 0 | 1 | 0 | 2 | 0 | 3.2521133 | 1.8642826 | 1.3878307 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | NOT_AVAILABLE | 207.04190534382894 | -16.76473078701521 | 84.77234739467015 | -25.880154554757564 | False | False | 0 | False | False | False | False | False | False | False | False | 8.234745e-13 | 1.9977706e-09 | 0.9999949 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | 0.08287548585527736 |
reduced_query = gaia_query['ra', 'dec', 'phot_g_mean_mag']
reduced_query.write('gaia_no_pm.cat', format='ascii.commented_header', overwrite=True)
reduced_query
| ra | dec | phot_g_mean_mag |
|---|---|---|
| deg | deg | mag |
| float64 | float64 | float32 |
| 85.25078206874738 | -2.444257621037541 | 19.836756 |
| 85.24865858921503 | -2.441354584616136 | 17.722948 |
| 85.25474417063919 | -2.470084090608095 | 17.202978 |
| 85.26833524857398 | -2.4416029853336667 | 20.263578 |
| 85.2386085747217 | -2.462292300986681 | 20.337946 |
| 85.23467309871081 | -2.4498085573057633 | 20.23374 |
| 85.27075582329469 | -2.441307234608435 | 19.156166 |
| 85.25578005166672 | -2.4765054474694934 | 20.352 |
| 85.27451682697576 | -2.472305714228985 | 20.950966 |
| ... | ... | ... |
| 85.31116982575814 | -2.5106681948456453 | 19.762535 |
| 85.18852621317357 | -2.405656267543347 | 19.284597 |
| 85.19030586577739 | -2.5028819323642013 | 17.53799 |
| 85.17292618358479 | -2.4623957493034743 | 19.089468 |
| 85.30495708910514 | -2.5178904392586623 | 20.880104 |
| 85.33205874915119 | -2.425249860759321 | 20.068647 |
| 85.30747238821475 | -2.38949359231377 | 19.625616 |
| 85.24559042309376 | -2.535389226550673 | 20.522165 |
| 85.29328038861492 | -2.526244532656098 | 19.140984 |
2.2 Create a catalog of Gaia DR3 sources with Proper Motion Data #
This method uses the image FLT footprints and gives 161 sources, compared to 183 with the prior method.
pm_cat = create_astrometric_catalog(sorted(glob.glob('*flt.fits')))
pm_cat.write('gaia_pm.cat', overwrite=True, format='ascii.no_header')
len(pm_cat)
2025336202243 INFO src=drizzlepac.haputils.astrometric_utils- Getting catalog using:
http://gsss.stsci.edu/webservices/vo/CatalogSearch.aspx?RA=85.25358125611297&DEC=-2.4517874752980333&SR=0.07776447588540084&FORMAT=CSV&CAT=GAIAedr3&MINDET=5&EPOCH=2012.839
2025336202244 INFO src=drizzlepac.haputils.astrometric_utils- Created catalog 'ref_cat.ecsv' with 161 sources
161
2.3 Run TweakReg #
Next we run TweakReg on the visit-level drizzled (DRZ) images and align to the Gaia catalog with proper motion data.
Because the fit RMS values for the MAST products were large for some visits, we allow for a larger than usual search radius of 1”. We also set the conv_width value slightly higher than the recommended value of 2.5 for WFC3/IR data in order to use barely resolved sources for alignment.
refcat = 'gaia_pm.cat' # Use the catalog with proper motion data
drz_files = sorted(glob.glob('*drz.fits'))
tweakreg.TweakReg(drz_files,
imagefindcfg={'threshold': 4, 'conv_width': 4.5},
minobj=3,
shiftfile=True,
outshifts='shift_drz.txt',
refcat=refcat,
searchrad=1,
ylimit=0.4,
nclip=1,
updatehdr=False, # change later when you verify the alignment works
interactive=False)
# clear_output()
Setting up logfile : tweakreg.log
TweakReg Version 3.10.0 started at: 20:22:44.575 (02/12/2025)
Version Information
--------------------
Python Version 3.11.14 | packaged by conda-forge | (main, Oct 22 2025, 22:46:25) [GCC 14.3.0]
numpy Version -> 2.3.5
astropy Version -> 7.2.0
stwcs Version -> 1.7.5
photutils Version -> 2.3.0
Finding shifts for:
ibxl50030_drz.fits
ibxl51030_drz.fits
ibxl52030_drz.fits
ibxl53030_drz.fits
ibxl54030_drz.fits
ibxl55030_drz.fits
ibxl56030_drz.fits
ibxl57030_drz.fits
ibxl58030_drz.fits
=== Source finding for image 'ibxl50030_drz.fits':
# Source finding for 'ibxl50030_drz.fits', EXT=('SCI', 1) started at: 20:22:44.618 (02/12/2025)
Found 75 objects.
=== FINAL number of objects in image 'ibxl50030_drz.fits': 75
=== Source finding for image 'ibxl51030_drz.fits':
# Source finding for 'ibxl51030_drz.fits', EXT=('SCI', 1) started at: 20:22:44.712 (02/12/2025)
Found 83 objects.
=== FINAL number of objects in image 'ibxl51030_drz.fits': 83
=== Source finding for image 'ibxl52030_drz.fits':
# Source finding for 'ibxl52030_drz.fits', EXT=('SCI', 1) started at: 20:22:44.803 (02/12/2025)
Found 87 objects.
=== FINAL number of objects in image 'ibxl52030_drz.fits': 87
=== Source finding for image 'ibxl53030_drz.fits':
# Source finding for 'ibxl53030_drz.fits', EXT=('SCI', 1) started at: 20:22:44.892 (02/12/2025)
Found 51 objects.
=== FINAL number of objects in image 'ibxl53030_drz.fits': 51
=== Source finding for image 'ibxl54030_drz.fits':
# Source finding for 'ibxl54030_drz.fits', EXT=('SCI', 1) started at: 20:22:44.97 (02/12/2025)
Found 54 objects.
=== FINAL number of objects in image 'ibxl54030_drz.fits': 54
=== Source finding for image 'ibxl55030_drz.fits':
# Source finding for 'ibxl55030_drz.fits', EXT=('SCI', 1) started at: 20:22:45.067 (02/12/2025)
Found 99 objects.
=== FINAL number of objects in image 'ibxl55030_drz.fits': 99
=== Source finding for image 'ibxl56030_drz.fits':
# Source finding for 'ibxl56030_drz.fits', EXT=('SCI', 1) started at: 20:22:45.157 (02/12/2025)
Found 73 objects.
=== FINAL number of objects in image 'ibxl56030_drz.fits': 73
=== Source finding for image 'ibxl57030_drz.fits':
# Source finding for 'ibxl57030_drz.fits', EXT=('SCI', 1) started at: 20:22:45.245 (02/12/2025)
Found 93 objects.
=== FINAL number of objects in image 'ibxl57030_drz.fits': 93
=== Source finding for image 'ibxl58030_drz.fits':
# Source finding for 'ibxl58030_drz.fits', EXT=('SCI', 1) started at: 20:22:45.334 (02/12/2025)
Found 103 objects.
=== FINAL number of objects in image 'ibxl58030_drz.fits': 103
===============================================================
Performing alignment in the projection plane defined by the WCS
derived from 'ibxl50030_drz.fits'
===============================================================
====================
Performing fit for: ibxl50030_drz.fits
Matching sources from 'ibxl50030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, -0.01949 with significance of 6.702 and 9 matches
Found 9 matches for ibxl50030_drz.fits...
Computed rscale fit for ibxl50030_drz.fits :
XSH: -0.9148 YSH: -0.4434 ROT: 0.022802346 SCALE: 1.000451
FIT XRMS: 0.051 FIT YRMS: 0.082
FIT RMSE: 0.097 FIT MAE: 0.077
RMS_RA: 2.3e-06 (deg) RMS_DEC: 2.6e-06 (deg)
Final solution based on 9 objects.
wrote XY data to: ibxl50030_drz_catalog_fit.match
Total # points: 9
# of points after clipping: 9
Total # points: 9
# of points after clipping: 9
====================
Performing fit for: ibxl51030_drz.fits
Matching sources from 'ibxl51030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of -0.1306, -0.2973 with significance of 3.909 and 12 matches
Found 12 matches for ibxl51030_drz.fits...
Computed rscale fit for ibxl51030_drz.fits :
XSH: -0.0950 YSH: -0.6794 ROT: 359.9831545 SCALE: 1.000220
FIT XRMS: 0.039 FIT YRMS: 0.043
FIT RMSE: 0.058 FIT MAE: 0.056
RMS_RA: 1.6e-06 (deg) RMS_DEC: 1.3e-06 (deg)
Final solution based on 12 objects.
wrote XY data to: ibxl51030_drz_catalog_fit.match
Total # points: 12
# of points after clipping: 12
Total # points: 12
# of points after clipping: 12
====================
Performing fit for: ibxl52030_drz.fits
Matching sources from 'ibxl52030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, -0.6723 with significance of 6.702 and 8 matches
Found 8 matches for ibxl52030_drz.fits...
Computed rscale fit for ibxl52030_drz.fits :
XSH: -0.5099 YSH: -0.3491 ROT: 0.001210839284 SCALE: 0.999709
FIT XRMS: 0.038 FIT YRMS: 0.018
FIT RMSE: 0.042 FIT MAE: 0.037
RMS_RA: 1.4e-06 (deg) RMS_DEC: 4.3e-07 (deg)
Final solution based on 8 objects.
wrote XY data to: ibxl52030_drz_catalog_fit.match
Total # points: 8
# of points after clipping: 8
Total # points: 8
# of points after clipping: 8
====================
Performing fit for: ibxl53030_drz.fits
Matching sources from 'ibxl53030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, -0.7973 with significance of 5 and 5 matches
Found 5 matches for ibxl53030_drz.fits...
Computed rscale fit for ibxl53030_drz.fits :
XSH: -0.2544 YSH: -0.8531 ROT: 359.9968781 SCALE: 1.000185
FIT XRMS: 0.052 FIT YRMS: 0.053
FIT RMSE: 0.074 FIT MAE: 0.062
RMS_RA: 2.1e-06 (deg) RMS_DEC: 1.6e-06 (deg)
Final solution based on 5 objects.
wrote XY data to: ibxl53030_drz_catalog_fit.match
Total # points: 5
# of points after clipping: 5
Total # points: 5
# of points after clipping: 5
====================
Performing fit for: ibxl54030_drz.fits
Matching sources from 'ibxl54030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, -0.7973 with significance of 7 and 7 matches
Found 7 matches for ibxl54030_drz.fits...
Computed rscale fit for ibxl54030_drz.fits :
XSH: -0.1959 YSH: -0.9179 ROT: 359.9732889 SCALE: 0.999206
FIT XRMS: 0.033 FIT YRMS: 0.049
FIT RMSE: 0.059 FIT MAE: 0.049
RMS_RA: 1.4e-06 (deg) RMS_DEC: 1.5e-06 (deg)
Final solution based on 7 objects.
wrote XY data to: ibxl54030_drz_catalog_fit.match
Total # points: 7
# of points after clipping: 7
Total # points: 7
# of points after clipping: 7
====================
Performing fit for: ibxl55030_drz.fits
Matching sources from 'ibxl55030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.02091, -0.4336 with significance of 5.326 and 11 matches
Found 10 matches for ibxl55030_drz.fits...
Computed rscale fit for ibxl55030_drz.fits :
XSH: -0.0106 YSH: -0.4748 ROT: 359.9953351 SCALE: 1.000175
FIT XRMS: 0.054 FIT YRMS: 0.039
FIT RMSE: 0.066 FIT MAE: 0.06
RMS_RA: 2.1e-06 (deg) RMS_DEC: 1.1e-06 (deg)
Final solution based on 10 objects.
wrote XY data to: ibxl55030_drz_catalog_fit.match
Total # points: 10
# of points after clipping: 10
Total # points: 10
# of points after clipping: 10
====================
Performing fit for: ibxl56030_drz.fits
Matching sources from 'ibxl56030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, -0.7973 with significance of 5 and 5 matches
Found 5 matches for ibxl56030_drz.fits...
Computed rscale fit for ibxl56030_drz.fits :
XSH: -0.1540 YSH: -0.5500 ROT: 359.9989578 SCALE: 1.000177
FIT XRMS: 0.14 FIT YRMS: 0.065
FIT RMSE: 0.15 FIT MAE: 0.12
RMS_RA: 5.1e-06 (deg) RMS_DEC: 1.5e-06 (deg)
Final solution based on 5 objects.
wrote XY data to: ibxl56030_drz_catalog_fit.match
Total # points: 5
# of points after clipping: 5
Total # points: 5
# of points after clipping: 5
====================
Performing fit for: ibxl57030_drz.fits
Matching sources from 'ibxl57030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.2027, 0.2027 with significance of 12 and 12 matches
Found 12 matches for ibxl57030_drz.fits...
Computed rscale fit for ibxl57030_drz.fits :
XSH: 0.0105 YSH: 0.0838 ROT: 0.001620903605 SCALE: 1.000051
FIT XRMS: 0.038 FIT YRMS: 0.052
FIT RMSE: 0.064 FIT MAE: 0.055
RMS_RA: 1.6e-06 (deg) RMS_DEC: 1.6e-06 (deg)
Final solution based on 12 objects.
wrote XY data to: ibxl57030_drz_catalog_fit.match
Total # points: 12
# of points after clipping: 12
Total # points: 12
# of points after clipping: 12
====================
Performing fit for: ibxl58030_drz.fits
Matching sources from 'ibxl58030_drz.fits' with sources from reference catalog 'gaia_pm.cat'
Computing initial guess for X and Y shifts...
Found initial X and Y shifts of 0.0694, 0.2027 with significance of 12.52 and 15 matches
Found 14 matches for ibxl58030_drz.fits...
Computed rscale fit for ibxl58030_drz.fits :
XSH: 0.0134 YSH: 0.0520 ROT: 359.998648 SCALE: 0.999998
FIT XRMS: 0.073 FIT YRMS: 0.051
FIT RMSE: 0.089 FIT MAE: 0.072
RMS_RA: 2.8e-06 (deg) RMS_DEC: 1.4e-06 (deg)
Final solution based on 14 objects.
wrote XY data to: ibxl58030_drz_catalog_fit.match
Total # points: 14
# of points after clipping: 14
Total # points: 14
# of points after clipping: 14
Writing out shiftfile : shift_drz.txt
Trailer file written to: tweakreg.log
# If the alignment is unsuccessful, stop the notebook
with open('shift_drz.txt', 'r') as shift:
for line_number, line in enumerate(shift, start=1):
if "nan" in line:
raise ValueError(f'nan found in line {line_number} in shift file')
2.4 Inspect the shift file and fit quality #
# Read the shift file just created by tweakreg
# There are 7 columns including: filename, x-shift, y-shift, rotation, scale, x-RMS, and y-RMS
shift_table = Table.read('shift_drz.txt',
format='ascii.no_header',
names=['file', 'dx', 'dy', 'rot', 'scale', 'xrms', 'yrms'])
# Define the format for each column (excluding 'file').
formats = ['.2f', '.2f', '.3f', '.5f', '.2f', '.2f']
# Iterate over the columns 'dx', 'dy', 'rot', 'scale', 'xrms', 'yrms'
for i, col in enumerate(shift_table.colnames[1:]):
# Apply the format to the current column
shift_table[col].format = formats[i]
# Display the table in the notebook
shift_table
| file | dx | dy | rot | scale | xrms | yrms |
|---|---|---|---|---|---|---|
| str18 | float64 | float64 | float64 | float64 | float64 | float64 |
| ibxl50030_drz.fits | -0.91 | -0.44 | 0.023 | 1.00045 | 0.05 | 0.08 |
| ibxl51030_drz.fits | -0.10 | -0.68 | 359.983 | 1.00022 | 0.04 | 0.04 |
| ibxl52030_drz.fits | -0.51 | -0.35 | 0.001 | 0.99971 | 0.04 | 0.02 |
| ibxl53030_drz.fits | -0.25 | -0.85 | 359.997 | 1.00019 | 0.05 | 0.05 |
| ibxl54030_drz.fits | -0.20 | -0.92 | 359.973 | 0.99921 | 0.03 | 0.05 |
| ibxl55030_drz.fits | -0.01 | -0.47 | 359.995 | 1.00018 | 0.05 | 0.04 |
| ibxl56030_drz.fits | -0.15 | -0.55 | 359.999 | 1.00018 | 0.14 | 0.06 |
| ibxl57030_drz.fits | 0.01 | 0.08 | 0.002 | 1.00005 | 0.04 | 0.05 |
| ibxl58030_drz.fits | 0.01 | 0.05 | 359.999 | 1.00000 | 0.07 | 0.05 |
Note that there are large residual rotation and scale terms in the shift file for several visits in the MAST data products. These will be corrected when we run TweakReg an additional time and update the WCS in Section 2.6 below.
match_files = sorted(glob.glob('*_drz_catalog_fit.match'))
for f in match_files:
input = ascii.read(f)
print(f'Number of matches for {f} {len(input)}')
Number of matches for ibxl50030_drz_catalog_fit.match 9
Number of matches for ibxl51030_drz_catalog_fit.match 12
Number of matches for ibxl52030_drz_catalog_fit.match 8
Number of matches for ibxl53030_drz_catalog_fit.match 5
Number of matches for ibxl54030_drz_catalog_fit.match 7
Number of matches for ibxl55030_drz_catalog_fit.match 10
Number of matches for ibxl56030_drz_catalog_fit.match 5
Number of matches for ibxl57030_drz_catalog_fit.match 12
Number of matches for ibxl58030_drz_catalog_fit.match 14
Next we compare with the MAST WCS solutions, number of matches and fit RMS values. Since there are only 5 Gaia sources in Visits 53 and 56, these did not have a successful Gaia fit during MAST processing and instead were aligned to the next catalog in the priority list, GSC v2.4.2. (Currently the minimum number of matches for a successful fit is 6, but this will updated to 10 in summer 2024.
wcstable
| FILENAME | DETECTOR | WCSNAME | NMATCHES | RMS_RA | RMS_DEC | RMS_RA_pix | RMS_DEC_pix |
|---|---|---|---|---|---|---|---|
| str18 | str2 | str30 | int64 | float64 | float64 | float64 | float64 |
| ibxl50030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 18 | 66.6 | 52.5 | 0.52 | 0.41 |
| ibxl51030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 31 | 73.7 | 86.3 | 0.57 | 0.67 |
| ibxl52030_drz.fits | IR | IDC_w3m18525i-FIT_IMG_GSC242 | 20 | 58.5 | 66.1 | 0.46 | 0.51 |
| ibxl53030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 12 | 55.6 | 75.8 | 0.43 | 0.59 |
| ibxl54030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 14 | 72.8 | 88.0 | 0.57 | 0.69 |
| ibxl55030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 19 | 63.5 | 86.9 | 0.49 | 0.68 |
| ibxl56030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GSC242 | 18 | 61.9 | 93.4 | 0.48 | 0.73 |
| ibxl57030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GAIAeDR3 | 12 | 5.8 | 6.5 | 0.05 | 0.05 |
| ibxl58030_drz.fits | IR | IDC_w3m18525i-FIT_REL_GAIAeDR3 | 15 | 11.5 | 6.8 | 0.09 | 0.05 |
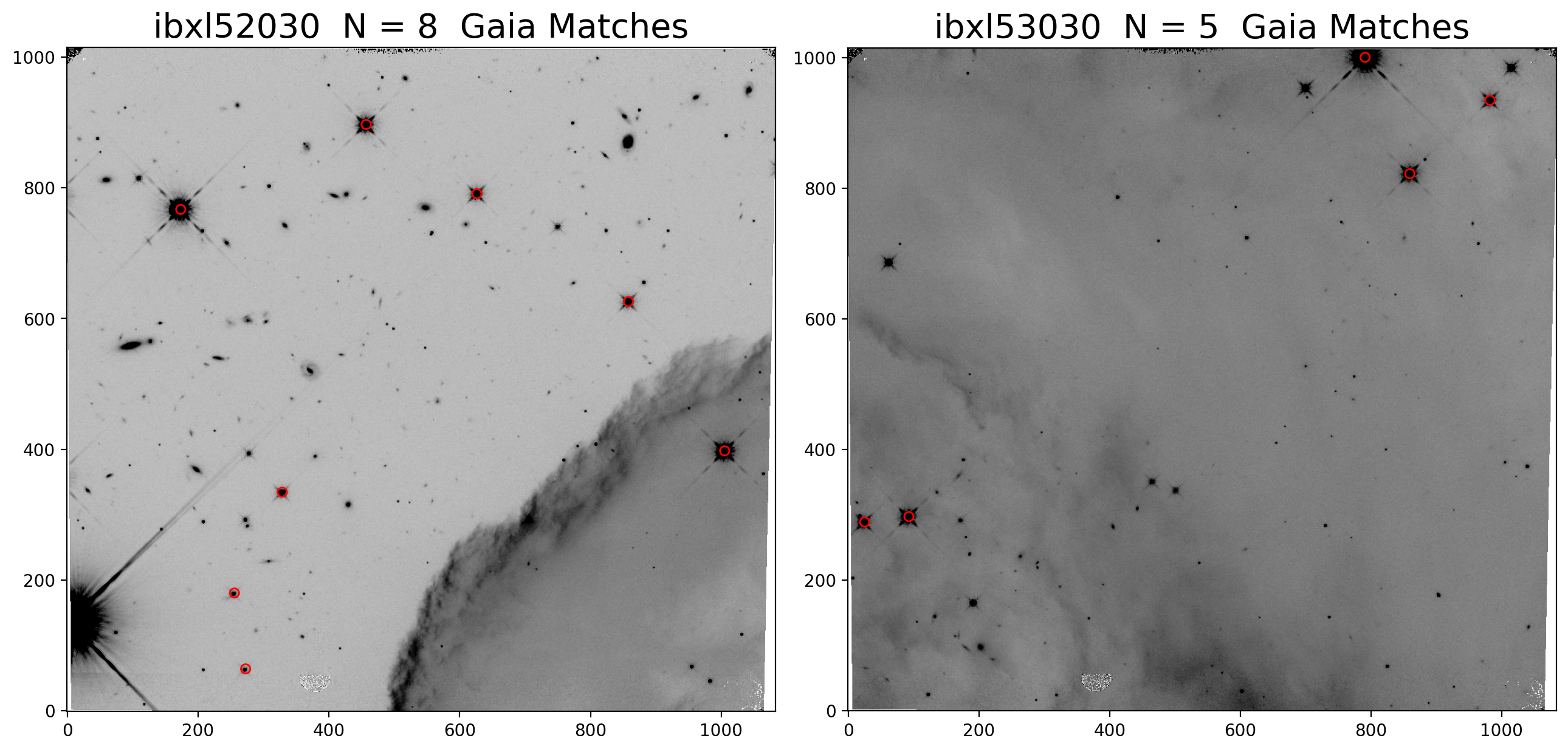
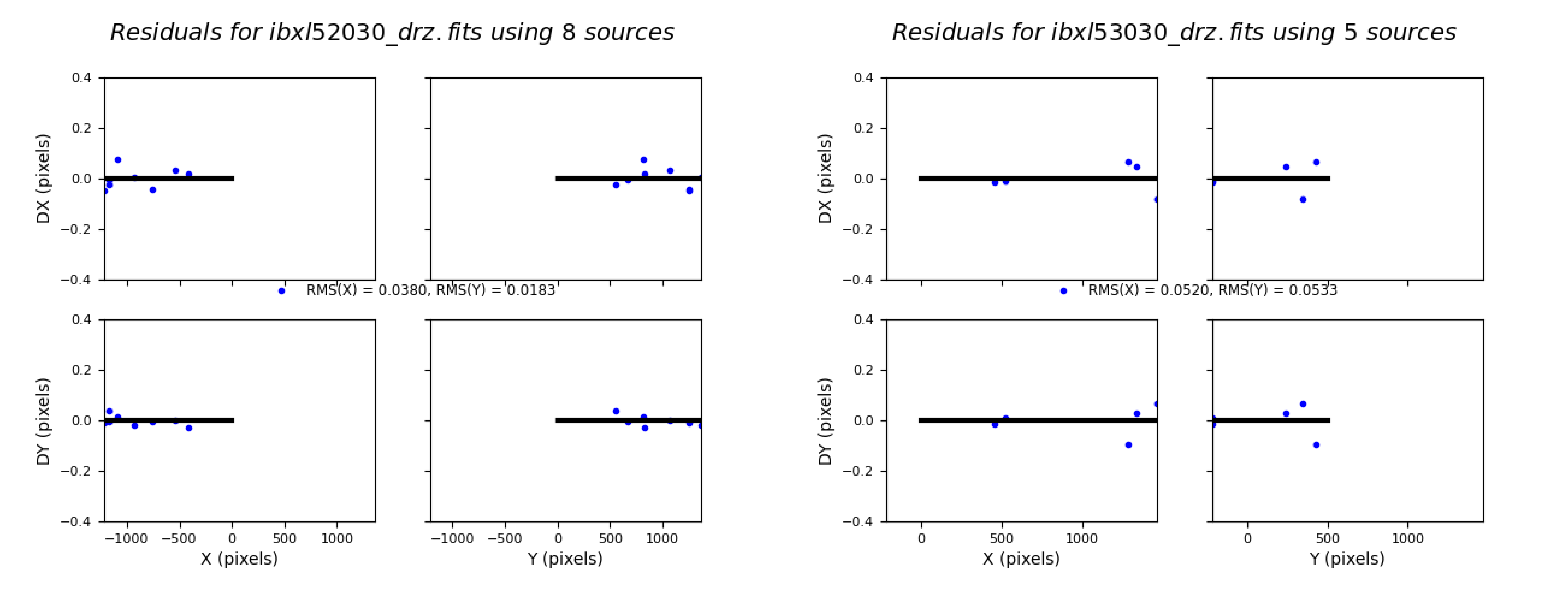
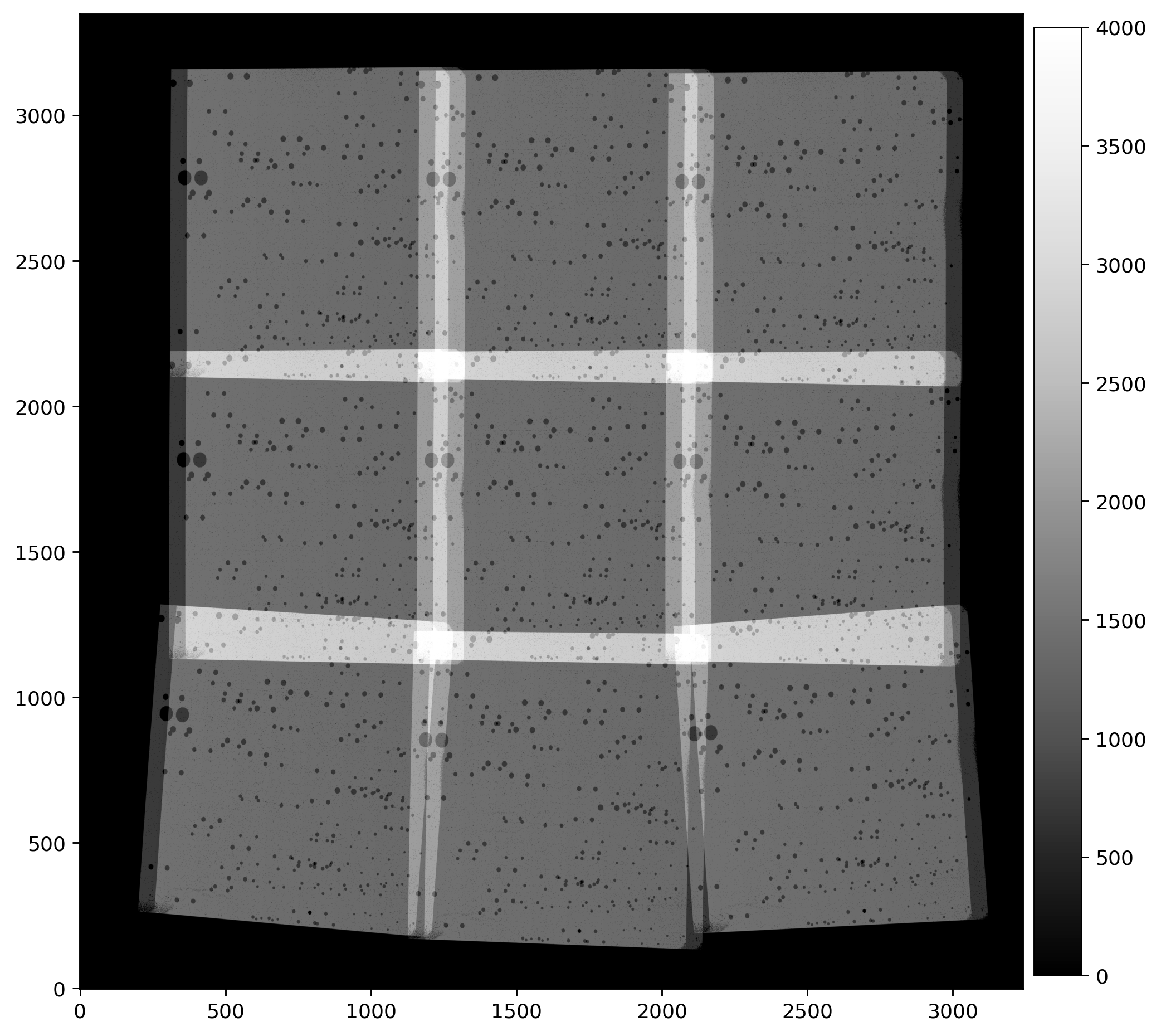
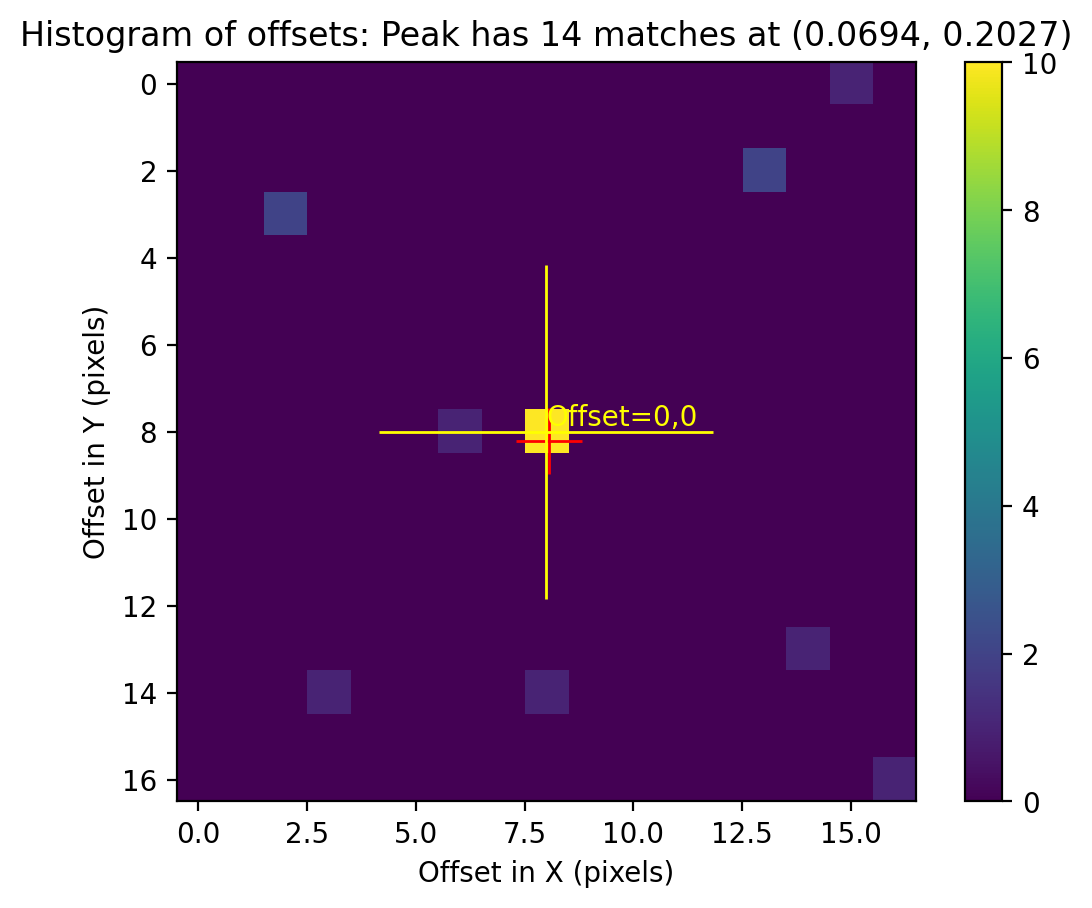
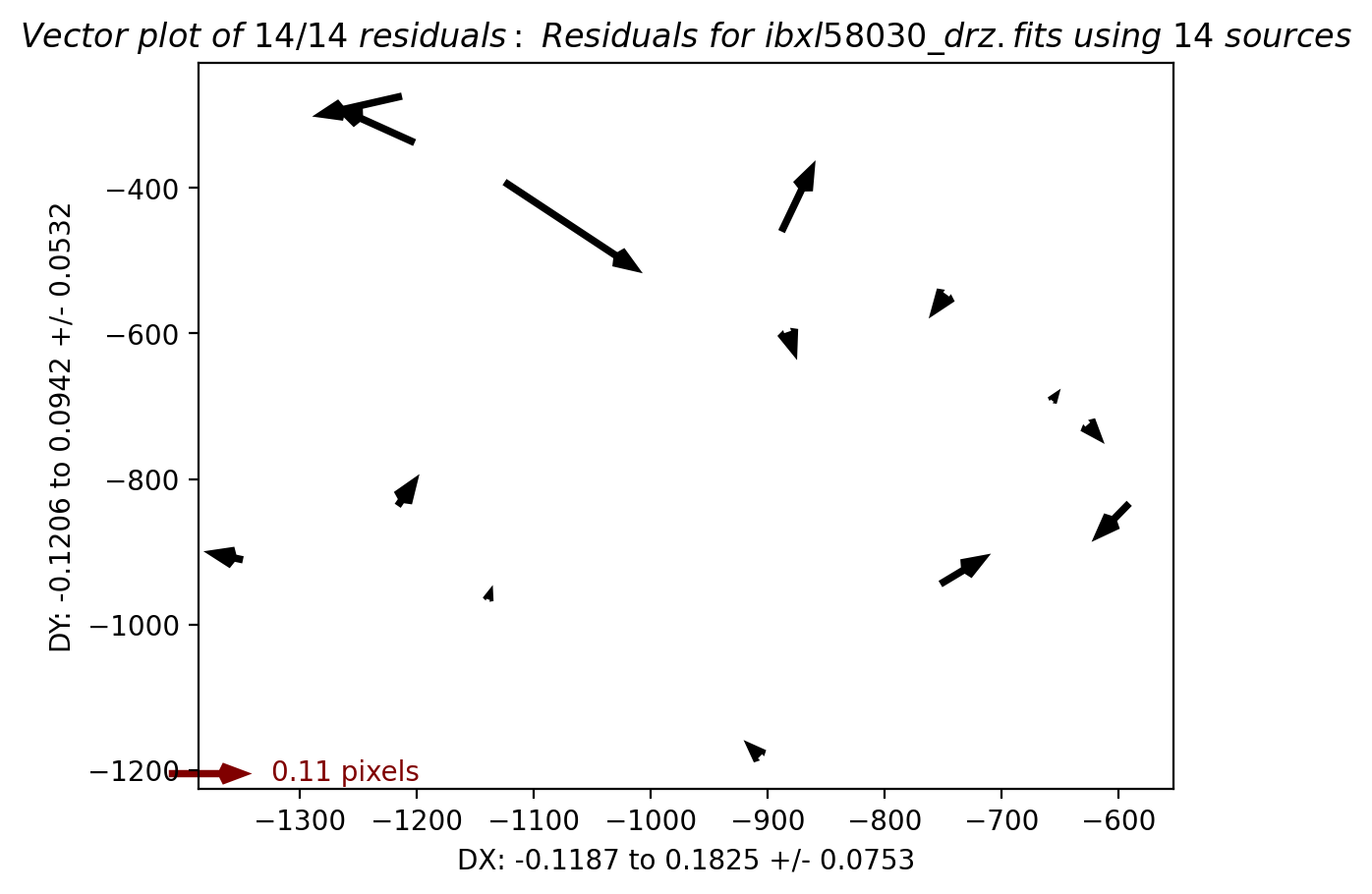
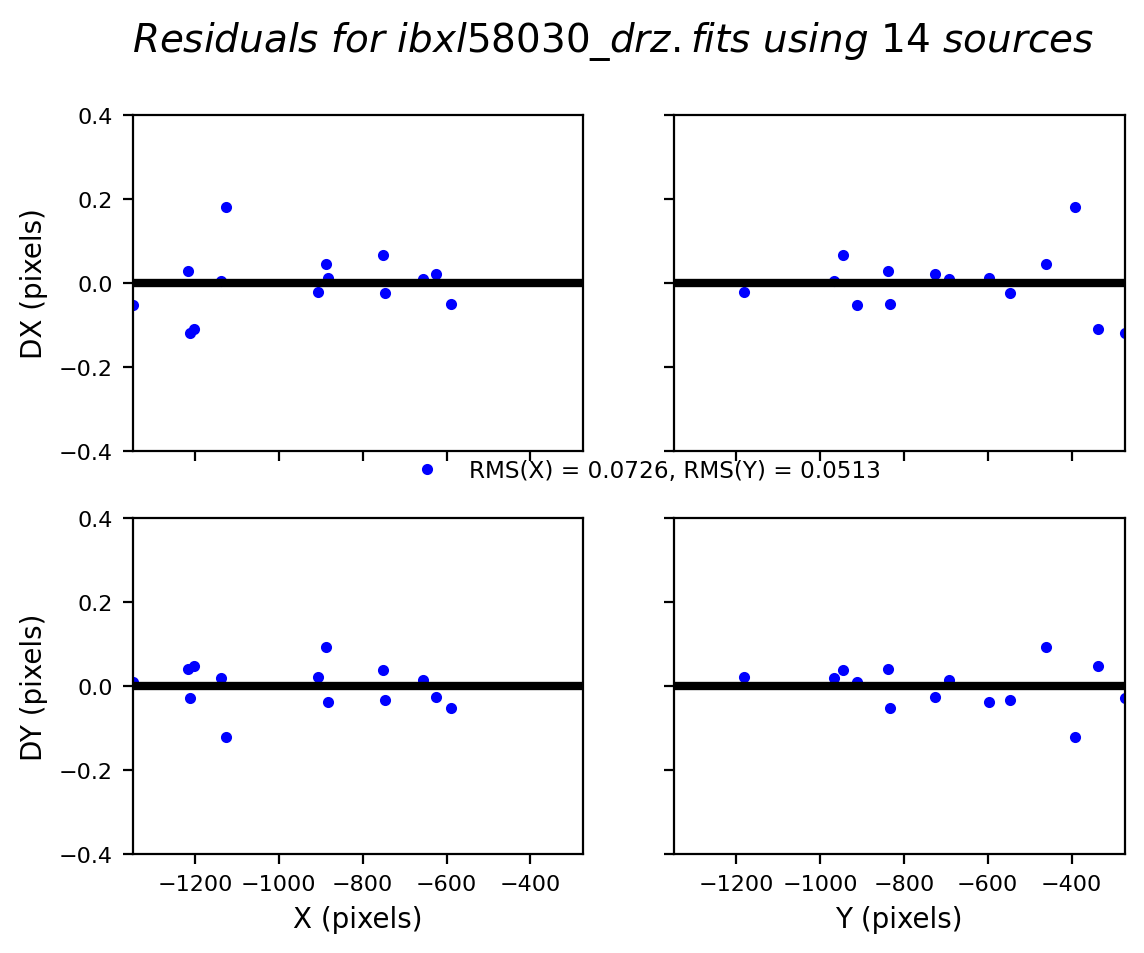
2.5 Overplot matched sources and inspect fit residuals #
Here we overplot the HST sources which were successfully matched with Gaia eDR3 and we look at the astrometric fit residual PNG plots.
While we inspect only two visits (52 and 53) at a time, and additional visits may be uncommented in the cell below.
# Define the rootnames for the two FITS files to be compared
# Uncomment the pairs you want to use
# rootname_A = 'ibxl50030'
# rootname_B = 'ibxl51030'
rootname_A = 'ibxl52030'
rootname_B = 'ibxl53030'
# rootname_A = 'ibxl54030'
# rootname_B = 'ibxl55030'
# rootname_A = 'ibxl56030'
# rootname_B = 'ibxl57030'
# rootname_A = 'ibxl57030'
# rootname_B = 'ibxl58030'
# Create subplots with 1 row and 2 columns
fig, [ax1, ax2] = plt.subplots(1, 2, figsize=(13, 10))
# Open the FITS files and extract the data from the 'SCI' extension
data_a = fits.open(rootname_A+'_drz.fits')['SCI', 1].data
data_b = fits.open(rootname_B+'_drz.fits')['SCI', 1].data
# Display the images in grayscale with a stretch from 0 to 2
ax1.imshow(data_a, cmap='Greys', origin='lower', vmin=0, vmax=2)
ax2.imshow(data_b, cmap='Greys', origin='lower', vmin=0, vmax=2)
# Read the matching catalog files for both rootnames
match_tab_a = ascii.read(rootname_A+'_drz_catalog_fit.match')
match_tab_b = ascii.read(rootname_B+'_drz_catalog_fit.match')
# Extract x and y coordinates from the matching catalogs
x_coord_a, y_coord_a = match_tab_a['col11'], match_tab_a['col12']
x_coord_b, y_coord_b = match_tab_b['col11'], match_tab_b['col12']
# Plot the coordinates on the images with red circles
ax1.scatter(x_coord_a, y_coord_a, s=30, edgecolor='r', facecolor='None')
ax2.scatter(x_coord_b, y_coord_b, s=30, edgecolor='r', facecolor='None')
# Set the titles for the subplots, including the number of matches
ax1.set_title(rootname_A+f' N = {len(match_tab_a)} Gaia Matches', fontsize=20)
ax2.set_title(rootname_B+f' N = {len(match_tab_b)} Gaia Matches', fontsize=20)
fig.tight_layout()
# Load, display, and inspect fit residual PNG files
img_A = mpimg.imread(f'residuals_{rootname_A}_drz.png')
img_B = mpimg.imread(f'residuals_{rootname_B}_drz.png')
# Create subplots for the residual images
fig, axs = plt.subplots(1, 2, figsize=(20, 10), dpi=200)
axs[0].imshow(img_A)
axs[1].imshow(img_B)
# Remove unnecessary axis from the residual images
axs[0].axis('off')
axs[1].axis('off')
# Adjust layout to minimize margins and display plots
fig.tight_layout()
plt.show()
2.6 Rerun TweakReg and update the header WCS #
Once we are happy with the alignment, we run TweakReg again with the same parameters but with the updatehdr=True.
refcat = 'gaia_pm.cat' # Use the catalog with proper motion data
tweakreg.TweakReg(drz_files,
imagefindcfg={'threshold': 4, 'conv_width': 4.5},
minobj=4,
shiftfile=False,
refcat=refcat,
searchrad=1,
ylimit=0.4,
nclip=1,
updatehdr=True, # update header
interactive=False)
clear_output()
2.7 Run TweakBack to propogate the WCS to the FLT files #
Finally, we run Tweakback on the aligned DRZ files to propogate the updated WCS information back to the FLT files.
for f in drz_files:
tb_input = f+'[sci,1]'
with fits.open(f) as hdu:
tweakback.apply_tweak(tb_input, orig_wcs_name=hdu[1].header['WCSNAME'])
clear_output()
3. Compare skymethod options in AstroDrizzle #
Now that the FLT files contain the updated WCS solutions, we explore different algorithms for estimating the sky.
3.1 skymethod = 'localmin' #
When using AstroDrizzle to compute the sky in each frame, ‘localmin’ will compute a common sky value for all chips of a given exposure, using the minimum sky value from all chips. This process is repeated for each input image. This algorithm is recommended when images are dominated by blank sky instead of extended, diffuse sources.
See readthedocs for more details on sky subtraction options.
In the command below, the aligned FLT frames are sky subtracted and drizzled together.
Because the WFC3/IR data products are already cleaned of cosmic rays during calwf3 processing, cosmic-ray rejection is turned off in AstroDrizzle by setting the parameters driz_separate, median, blot, and driz_cr to ‘False’. Note that final_bits=16 means only the stable hot pixels are treated as good.
sky = 'localmin'
astrodrizzle.AstroDrizzle('*flt.fits',
output='f160w_localmin',
preserve=False,
context=False,
skymethod=sky,
driz_separate=False, median=False, blot=False, driz_cr=False, # CR-rej turned off
final_bits='16',
final_wcs=True,
final_rot=257.)
clear_output()
3.2 skymethod = 'match' #
When skymethod is set to ‘match’, differences in sky values between images in common sky regions will be computed. Thus, sky values will be relative (delta) to the sky computed in one of the input images whose sky value will be set to and reported as 0. This setting “equalizes” sky values between the images in large mosaics.
This is the recommended setting for images containing diffuse sources (e.g., galaxies, nebulae) covering significant parts of the image.
sky = 'match'
astrodrizzle.AstroDrizzle('*flt.fits',
output='f160w_match',
preserve=False,
context=False,
skymethod=sky,
driz_separate=False, median=False, blot=False, driz_cr=False, # CRREJ=None
final_bits='16',
final_wcs=True,
final_rot=257.)
clear_output()
3.3 skymethod = 'globalmin+match' #
When skymethod is set to ‘globalmin+match’, AstroDrizzle will first find a minimum “global” sky value in all input images and then use the ‘match’ method to equalize sky values between images.
sky = 'globalmin+match'
astrodrizzle.AstroDrizzle('*flt.fits',
output='f160w_globalmin_match',
preserve=False,
context=False,
skymethod=sky,
driz_separate=False, median=False, blot=False, driz_cr=False, # CRREJ=None
final_bits='16',
final_wcs=True,
final_rot=257.)
clear_output()
3.4 skymethod = 'globalmin' #
When skymethod is set to ‘globalmin’, a common sky value will be computed for all exposures. AstroDrizzle will compute sky values for each chip/image extension, find the minimum sky value from all the exposures, and then subtract that minimum sky value from all chips in all images.
This method may be useful when input images already have matched background values.
sky = 'globalmin'
astrodrizzle.AstroDrizzle('*flt.fits',
output='f160w_globalmin',
preserve=False,
context=False,
skymethod=sky,
driz_separate=False, median=False, blot=False, driz_cr=False, # CRREJ=None
final_bits='16',
final_wcs=True,
final_rot=257.)
clear_output()
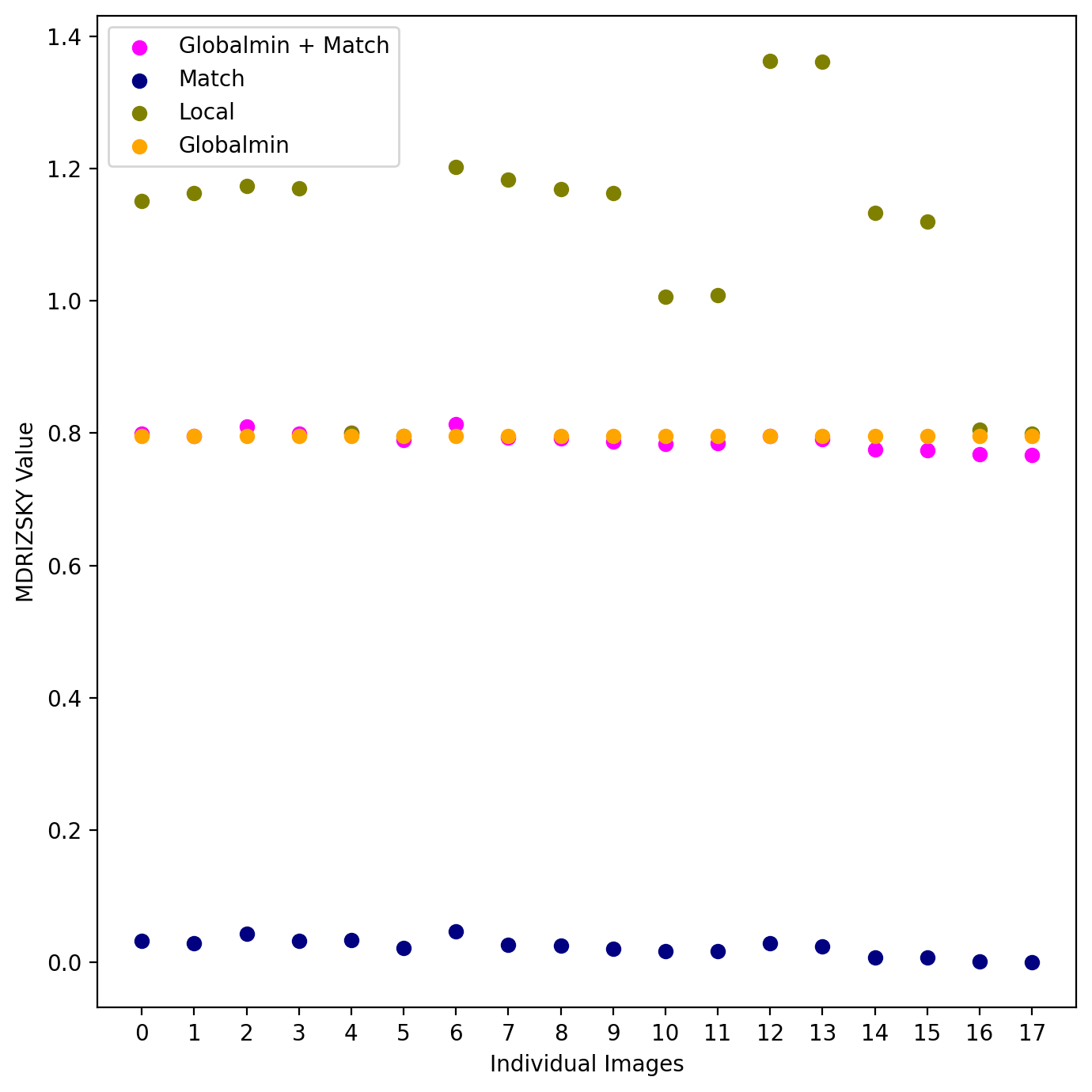
4. Compare the MDRIZSKY values for each method #
Below we provide a gif comparing the upper portion of the final drizzled image. We cycle through three drizzled images produced using different skymethod algorithms:
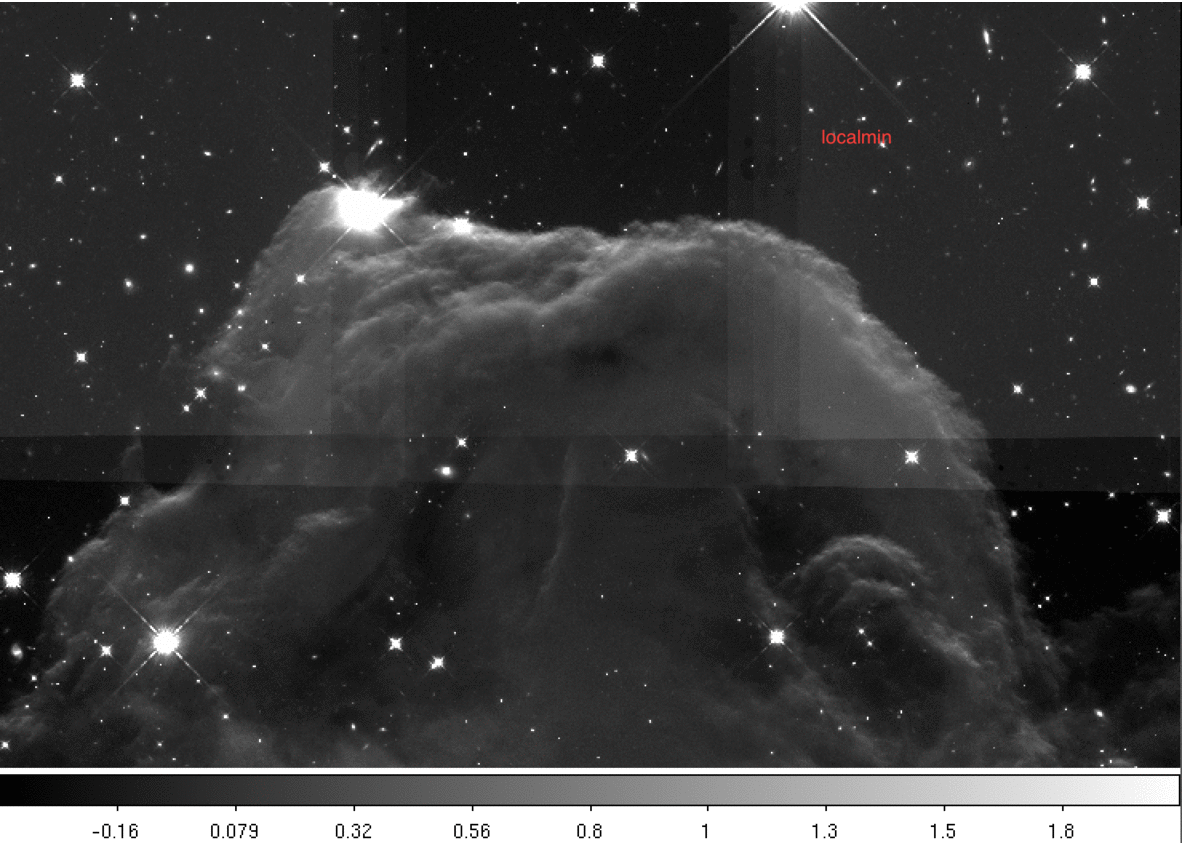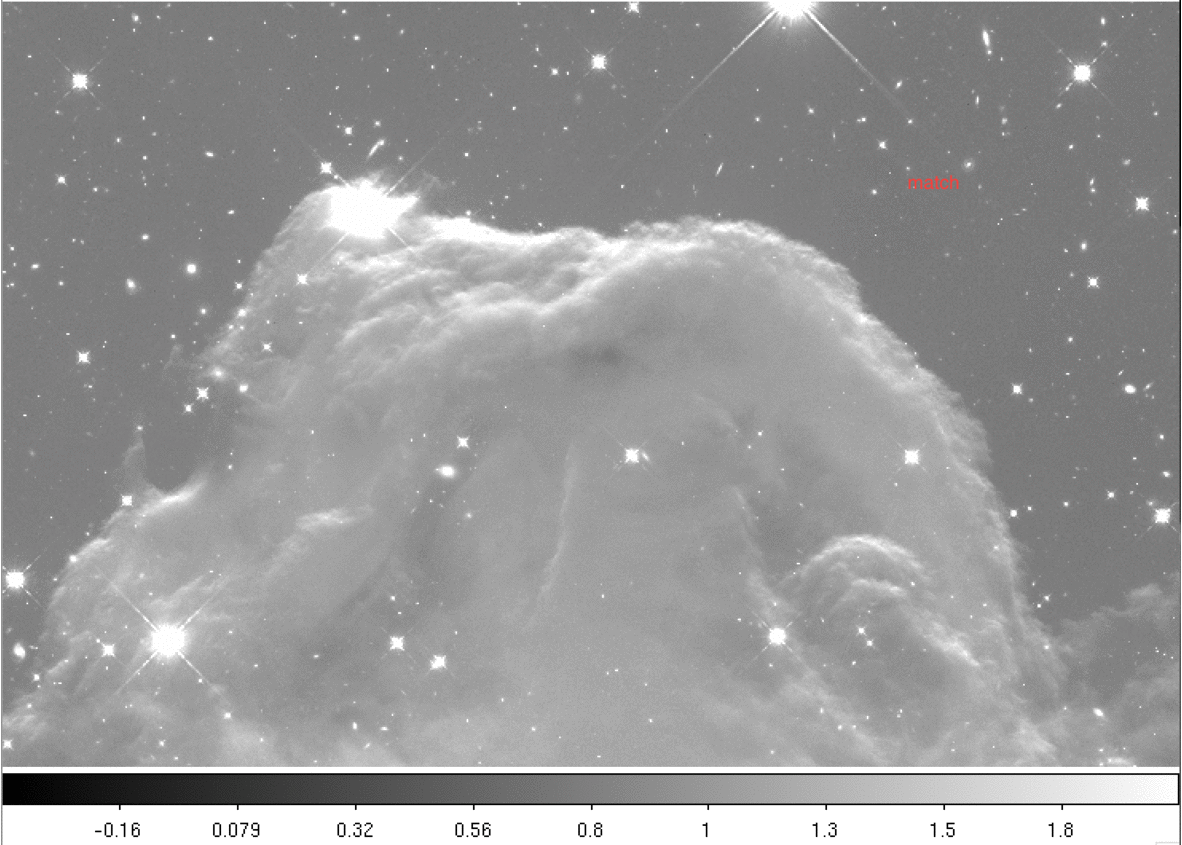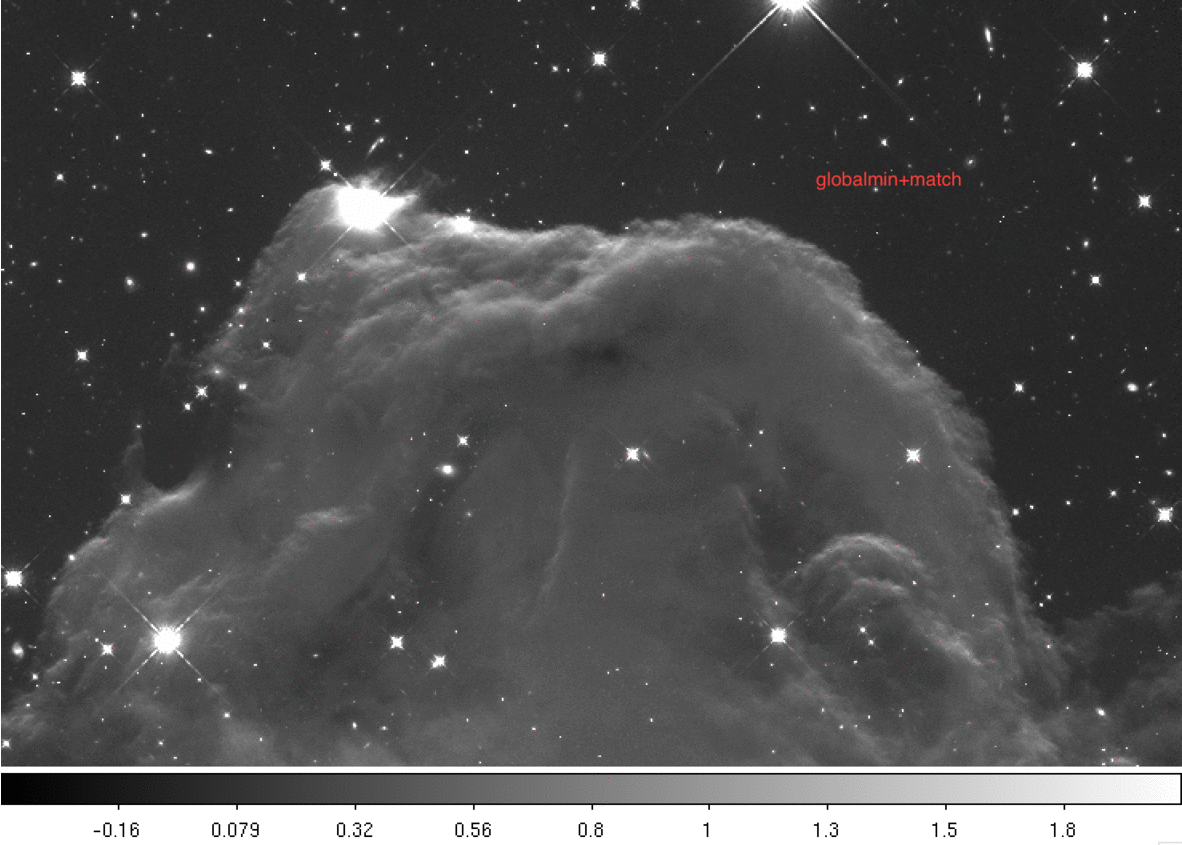
Next we print the sky values computed for each image using the four different methods.
mdrizsky_val = pandas.DataFrame({'rootname': fits.getdata('f160w_globalmin_drz_sci.fits', 1)['rootname'],
'local': fits.getdata('f160w_localmin_drz_sci.fits', 1)['mdrizsky'],
'globalmin': fits.getdata('f160w_globalmin_drz_sci.fits', 1)['mdrizsky'],
'globalmin_match': fits.getdata('f160w_globalmin_match_drz_sci.fits', 1)['mdrizsky'],
'match': fits.getdata('f160w_match_drz_sci.fits', 1)['mdrizsky']})
mdrizsky_val
| rootname | local | globalmin | globalmin_match | match | |
|---|---|---|---|---|---|
| 0 | ibxl50clq | 1.150338 | 0.795985 | 0.799595 | 0.032796 |
| 1 | ibxl50cqq | 1.162587 | 0.795985 | 0.795926 | 0.029128 |
| 2 | ibxl51eoq | 1.173177 | 0.795985 | 0.810412 | 0.043613 |
| 3 | ibxl51etq | 1.170005 | 0.795985 | 0.799362 | 0.032563 |
| 4 | ibxl52k0q | 0.801102 | 0.795985 | 0.801102 | 0.034303 |
| 5 | ibxl52k5q | 0.795985 | 0.795985 | 0.789244 | 0.022445 |
| 6 | ibxl53kxq | 1.202108 | 0.795985 | 0.813771 | 0.046972 |
| 7 | ibxl53l9q | 1.183192 | 0.795985 | 0.793237 | 0.026439 |
| 8 | ibxl54bgq | 1.169292 | 0.795985 | 0.792419 | 0.025620 |
| 9 | ibxl54blq | 1.162808 | 0.795985 | 0.787202 | 0.020403 |
| 10 | ibxl55f0q | 1.005760 | 0.795985 | 0.784272 | 0.017473 |
| 11 | ibxl55f5q | 1.008806 | 0.795985 | 0.784592 | 0.017793 |
| 12 | ibxl56huq | 1.362592 | 0.795985 | 0.796189 | 0.029390 |
| 13 | ibxl56i2q | 1.361517 | 0.795985 | 0.791313 | 0.024514 |
| 14 | ibxl57adq | 1.132586 | 0.795985 | 0.774923 | 0.008124 |
| 15 | ibxl57aiq | 1.119377 | 0.795985 | 0.774029 | 0.007230 |
| 16 | ibxl58sqq | 0.804761 | 0.795985 | 0.768317 | 0.001518 |
| 17 | ibxl58svq | 0.799282 | 0.795985 | 0.766799 | 0.000000 |
These computed sky values can be visualized in the plot below. To reiterate, the MDRIZSKY keyword reports the value subtracted from each FLC image prior to drizzling, and not the sky level itself. Thus the values for skymethod='match' are close to zero.
We also note that varying background levels across the individual tiles result in inaccurate sky background determination when skymethod='localmin' and thus a mismatched sky in the final mosaic.
index = mdrizsky_val.index.tolist()
globalmin = list(mdrizsky_val['globalmin'])
globalmin_match = list(mdrizsky_val['globalmin_match'])
match = list(mdrizsky_val['match'])
local = list(mdrizsky_val['local'])
# Plotting code:
fig = plt.figure(figsize=[7, 7])
plt.scatter(index, globalmin_match, color='magenta', label='Globalmin + Match')
plt.scatter(index, match, color='navy', label='Match')
plt.scatter(index, local, color='olive', label='Local')
plt.scatter(index, globalmin, color='orange', label='Globalmin')
plt.xlabel('Individual Images')
plt.ylabel('MDRIZSKY Value')
plt.legend(loc='best')
plt.xticks(index)
plt.tight_layout()
plt.show()
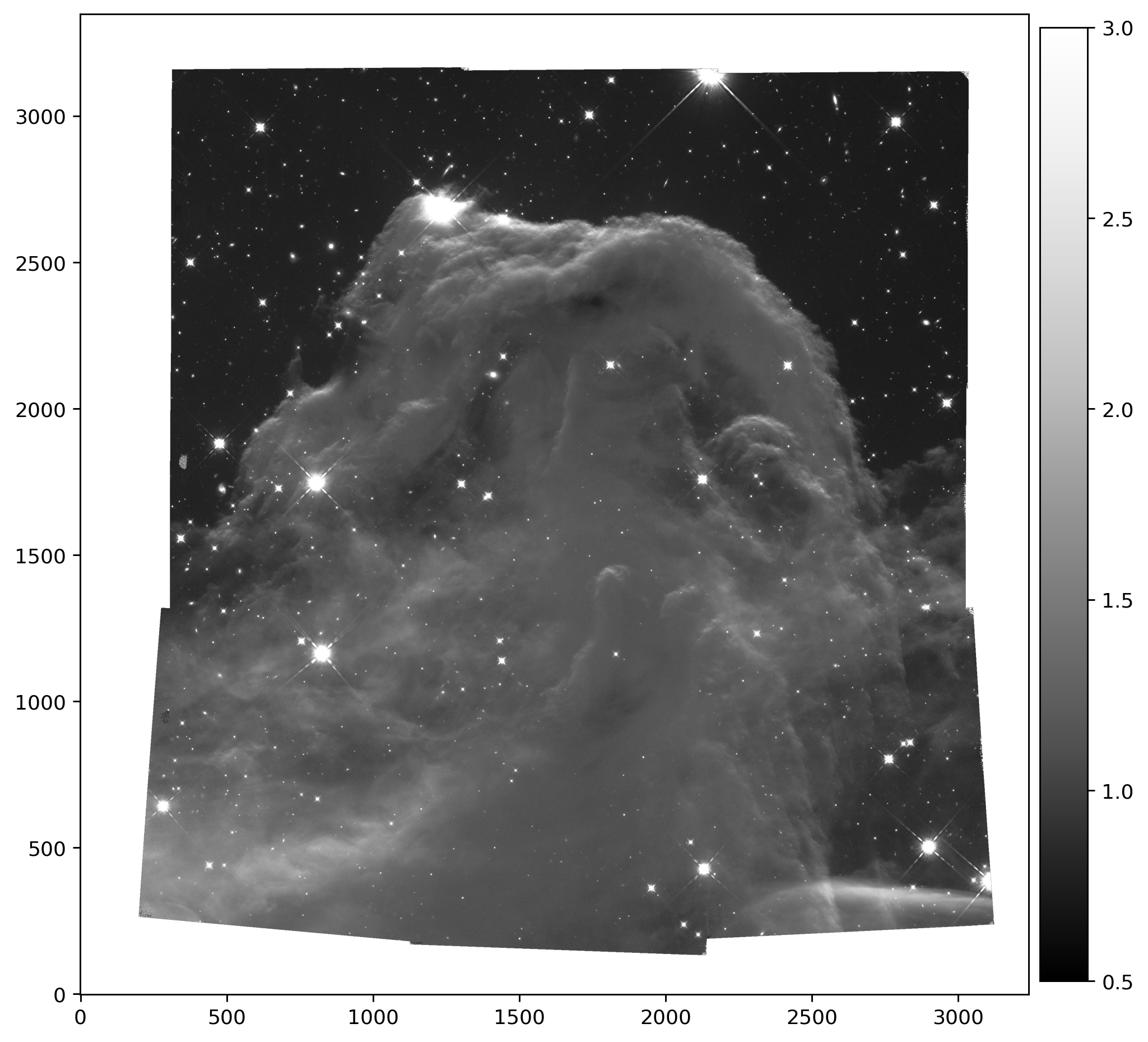
5. Display the ‘sky matched’ science mosaic and weight image #
Finally, we display the science and weight images for the combined mosaic.
sci = fits.getdata('f160w_match_drz_sci.fits')
fig = plt.figure(figsize=(10, 10), dpi=130)
plt.imshow(sci, vmin=0.5, vmax=3, cmap='Greys_r', origin='lower')
plt.colorbar(shrink=0.85, pad=0.01)
plt.show()
6. Conclusion #
Thank you for going through this notebook. You should now have all the necessary tools for assessing the
appropriate skymethod parameter to use when combining images. After completing this notebook you
should be more familiar with:
• How to effectively use astroquery to download FLT and DRZ files.
• Checking the WCS of images and aligning them to a reference catalog with TweakReg.
• Combining data with AstroDrizzle taking into account the skymethod
Congratulations, you have completed the notebook!
Additional Resources #
About this Notebook #
Created: 14 Dec 2018; C. Martlin & J. Mack
Updated: 16 Nov 2023; K. Huynh & J. Mack
Updated: 23 Jul 2024; J. Mack & B. Kuhn
Source: https://github.com/spacetelescope/hst_notebooks
Citations #
If you use Python packages for published research, please cite the authors. Follow these links for more
information about citing packages such as astropy, astroquery, matplotlib, pandas, etc.: