
Custom CCD Darks#
Table of Contents#
1. Introduction#
In the Calstis pipline for calibrating STIS CCD data, one of the steps is dark signal substraction, which removes the dark signal (count rate created in the detector in the absence of photons from the sky) from the uncalibrated science image based on reference file. Usually, the Calstis pipline uses the default dark reference file specified in the 0-extension header of the uncalibrated science image fits file. But for some faint sources, it is necessary to customize the dark reference for the observations to remove the hot pixels in the science image. In this notebook, we will go through how to create dark reference files using the Python refstis library.
Notice: this notebook demonstrates a squence of steps to customize dark reference file. If any of the intermediate steps fail or need to rerun, please restart the ipython kernel and start from the first step.
1.1 Import Necessary Packages#
astropy.io fitsastropy.tablefor accessing FITS filesastroquery.mast Observationsfor finding and downloading data from the MAST archiveos,shutil,pathlibfor managing system pathsnumpyto handle array functionsstistoolsfor calibrating STIS datarefstisfor creating STIS reference filesmatplotlibfor plotting data
For more information on installing refstis, see: Refstis: Superdarks and Superbiases for STIS
# Import for: Reading in fits file
from astropy.io import fits
from astropy.table import Table
# Import for: Downloading necessary files. (Not necessary if you choose to collect data from MAST)
from astroquery.mast import Observations
# Import for: Managing system variables and paths
import os
import shutil
from pathlib import Path
# Import for: Quick Calculation and Data Analysis
import numpy as np
# Import for: Operations on STIS Data
import stistools
from refstis.basedark import make_basedark
from refstis.weekdark import make_weekdark
# Import for: Plotting and specifying plotting parameters
import matplotlib
%matplotlib inline
from matplotlib import pyplot as plt
matplotlib.rcParams['image.origin'] = 'lower'
matplotlib.rcParams['image.cmap'] = 'plasma'
matplotlib.rcParams['image.interpolation'] = 'none'
matplotlib.rcParams['figure.figsize'] = (20, 10)
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/nmpfit.py:8: UserWarning: NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.
warnings.warn("NMPFIT is deprecated - stsci.tools v 3.5 is the last version to contain it.")
/home/runner/micromamba/envs/ci-env/lib/python3.11/site-packages/stsci/tools/gfit.py:18: UserWarning: GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it.Use astropy.modeling instead.
warnings.warn("GFIT is deprecated - stsci.tools v 3.4.12 is the last version to contain it."
The following tasks in the stistools package can be run with TEAL:
basic2d calstis ocrreject wavecal x1d x2d
1.2 Collect Data Set From the MAST Archive Using Astroquery#
There are other ways to download data from MAST such as using CyberDuck. The steps of collecting data is beyond the scope of this notebook, and we are only showing how to use astroquery and CRDS.
%%capture --no-display
# cleanup download directory
if os.path.exists('./mastDownload'):
shutil.rmtree('./mastDownload')
# change this field in you have a specific dataset to be explored
obs_id = "oeik1s030"
# Search target by obs_id
target = Observations.query_criteria(obs_id=obs_id)
# get a list of files assiciated with that target
FUV_list = Observations.get_product_list(target)
# Download fits files
result = Observations.download_products(FUV_list, extension='fits')
crj = os.path.join("./mastDownload/HST", "{}".format(obs_id), "{}_crj.fits".format(obs_id))
Next, use the Calibration Reference Data System (CRDS) command line tools to update and download the reference files for creating the basedark.
crds_path = os.path.expanduser("~") + "/crds_cache"
os.environ["CRDS_PATH"] = crds_path
os.environ["CRDS_SERVER_URL"] = "https://hst-crds.stsci.edu"
os.environ["oref"] = os.path.join(crds_path, "references/hst/oref/")
%%capture --no-stderr output
!crds bestrefs --update-bestrefs --sync-references=1 --files ./mastDownload/HST/oeik1s030/oeik1s030_raw.fits
2. Default Dark File#
The default dark file is specified in the 0th extension of an uncalibrated science image through a field called ‘DARKFILE’. We will later replace this default dark file with the customized dark file we created using refstis.
darkfile = fits.getval(crj, ext=0, keyword='DARKFILE')
print("The default dark file of observation {id} is: {df}".format(id=obs_id, df=darkfile))
The default dark file of observation oeik1s030 is: oref$55a20445o_drk.fits
3. Make Basedark#
Every month, a high signal-to-noise superdark frame is created from a combination of typically 40-60 “long” darks. These monthly superdark frames are not actually delivered to the calibration data base, but used as “baseline” dark for the next steps. When creating the Basedark, the input imsets are joined and combined into a single file, and the cosmic ray rejection is performed. Then the hot pixels in the combined image frame are identified and labeled in the DQ array using an iterative sigma clip method, and those hot pixels will later be updated with values in the Weekdark. In this section, we’ll show how to create the basedark file.
These superdark frames are not taken exactly each month, but during a roughly 30 days period called “annealing period”. The duration of each annealing period, together with the superdark frames taken, can be found here: STIS Annealing Periods.
We first get the observation date of our sample data, and find the corresponding anneal period:
TDATEOBS = fits.getval(crj, ext=0, keyword='TDATEOBS')
TTIMEOBS = fits.getval(crj, ext=0, keyword='TTIMEOBS')
print("UT date of start of first exposure in file is {}".format(TDATEOBS))
print("UT time of start of first exposure in file is {}".format(TTIMEOBS))
UT date of start of first exposure in file is 2021-05-05
UT time of start of first exposure in file is 12:29:54
According to the STIS Annealing Periods, this observation was taken during the annealing period from 2021-04-07 02:35:41 to 2021-05-05 14:00:22. We collect all the long component dark flt data during that annealing period:
%%capture --no-display
# copy the dark file obs_id from the STIS Annealing Periods table, and put them into a list
rootnames = "oeen8lqwq, oeen8ms5q, oeen8nvcq, oeen8oxnq, oeen8pa3q, oeen8qckq, oeen8reyq, oeen8sguq, "\
"oeen8taaq, oeen8udiq, oeen8vh3q, oeen8wicq, oeen8xkaq, oeen8yn4q, oeen8zpyq, oeen90rtq, oeen91u0q, oeen92wdq, "\
"oeen93ysq, oeen94arq, oeen95dlq, oeen96g4q, oeen97b7q, oeen98c4q, oeen99gxq, oeen9ah3q, oeen9bm7q, oeen9cmkq, "\
"oeen9dryq, oeen9et6q, oeen9fxwq, oeen9gy4q, oeen9icqq, oeen9hcwq, oeen9jgwq, oeen9kh4q, oeen9lafq, oeen9majq, "\
"oeen9nh9q, oeen9ohiq, oeen9qovq, oeen9ppcq, oeen9rucq, oeen9supq, oeen9tydq, oeen9uytq, oeen9velq, oeen9wf3q, "\
"oeen9xjeq, oeen9yjmq, oeen9za2q, oeena0aaq, oeena1g5q, oeena2gcq, oeena3kjq, oeena4knq".split(', ')
# search in astroquery based on obs_id
search = Observations.query_criteria(obs_id=rootnames)
pl = Observations.get_product_list(search)
# we only need the _flt fits files
pl = pl[pl['productSubGroupDescription'] == 'FLT']
# download the data
download_status = Observations.download_products(pl, mrp_only=False)
# store all the paths to the superdark frames into a list
anneal_dark = []
for root in rootnames:
file_path = os.path.join("./mastDownload/HST", "{}".format(root), "{}_flt.fits".format(root))
# check CCD amplifier
CCDAMP = fits.getval(file_path, keyword='CCDAMP', ext=0)
assert (CCDAMP == 'D')
anneal_dark.append(file_path)
filename_mapping = {os.path.basename(x).rsplit('_', 1)[0]: x for x in download_status['Local Path']}
Then we put the list of input dark files into refstis.make_basedark. The second parameter, refdark_name, is the name of the output basedark file. For detailed information on make_basedark, see: Basedark.
new_basedark = 'new_basedark.fits'
# remove the new_basefark file if it already exists
if os.path.exists(new_basedark):
os.remove(new_basedark)
make_basedark(anneal_dark, refdark_name=new_basedark, bias_file=None)
#-------------------------------#
# Running basedark #
#-------------------------------#
output to: new_basedark.fits
with biasfile None
./mastDownload/HST/oeen8lqwq/oeen8lqwq_flt.fits, ext 1: Scaling data by 0.928849220834831 for temperature: 19.0943
./mastDownload/HST/oeen8ms5q/oeen8ms5q_flt.fits, ext 1: Scaling data by 0.9506625167078937 for temperature: 18.7414
./mastDownload/HST/oeen8nvcq/oeen8nvcq_flt.fits, ext 1: Scaling data by 0.928849220834831 for temperature: 19.0943
./mastDownload/HST/oeen8oxnq/oeen8oxnq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen8pa3q/oeen8pa3q_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeen8qckq/oeen8qckq_flt.fits, ext 1: Scaling data by 0.8880994671403196 for temperature: 19.8
./mastDownload/HST/oeen8reyq/oeen8reyq_flt.fits, ext 1: Scaling data by 0.8880994671403196 for temperature: 19.8
./mastDownload/HST/oeen8sguq/oeen8sguq_flt.fits, ext 1: Scaling data by 0.8880994671403196 for temperature: 19.8
./mastDownload/HST/oeen8taaq/oeen8taaq_flt.fits, ext 1: Scaling data by 0.8880994671403196 for temperature: 19.8
./mastDownload/HST/oeen8udiq/oeen8udiq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen8vh3q/oeen8vh3q_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen8wicq/oeen8wicq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen8xkaq/oeen8xkaq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen8yn4q/oeen8yn4q_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen8zpyq/oeen8zpyq_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen90rtq/oeen90rtq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen91u0q/oeen91u0q_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen92wdq/oeen92wdq_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen93ysq/oeen93ysq_flt.fits, ext 1: Scaling data by 0.7915744812218742 for temperature: 21.7615
./mastDownload/HST/oeen94arq/oeen94arq_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen95dlq/oeen95dlq_flt.fits, ext 1: Scaling data by 0.7915744812218742 for temperature: 21.7615
./mastDownload/HST/oeen96g4q/oeen96g4q_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen97b7q/oeen97b7q_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen98c4q/oeen98c4q_flt.fits, ext 1: Scaling data by 0.7747337627424336 for temperature: 22.1538
./mastDownload/HST/oeen99gxq/oeen99gxq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen9ah3q/oeen9ah3q_flt.fits, ext 1: Scaling data by 0.8467959780578228 for temperature: 20.5846
./mastDownload/HST/oeen9bm7q/oeen9bm7q_flt.fits, ext 1: Scaling data by 0.8467959780578228 for temperature: 20.5846
./mastDownload/HST/oeen9cmkq/oeen9cmkq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9dryq/oeen9dryq_flt.fits, ext 1: Scaling data by 0.7585906599283587 for temperature: 22.5462
./mastDownload/HST/oeen9et6q/oeen9et6q_flt.fits, ext 1: Scaling data by 0.7915744812218742 for temperature: 21.7615
./mastDownload/HST/oeen9fxwq/oeen9fxwq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9gy4q/oeen9gy4q_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen9icqq/oeen9icqq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen9hcwq/oeen9hcwq_flt.fits, ext 1: Scaling data by 0.7915744812218742 for temperature: 21.7615
./mastDownload/HST/oeen9jgwq/oeen9jgwq_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeen9kh4q/oeen9kh4q_flt.fits, ext 1: Scaling data by 0.8880994671403196 for temperature: 19.8
./mastDownload/HST/oeen9lafq/oeen9lafq_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeen9majq/oeen9majq_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeen9nh9q/oeen9nh9q_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9ohiq/oeen9ohiq_flt.fits, ext 1: Scaling data by 0.8467959780578228 for temperature: 20.5846
./mastDownload/HST/oeen9qovq/oeen9qovq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9ppcq/oeen9ppcq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9rucq/oeen9rucq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9supq/oeen9supq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
./mastDownload/HST/oeen9tydq/oeen9tydq_flt.fits, ext 1: Scaling data by 0.8091636161198339 for temperature: 21.3692
./mastDownload/HST/oeen9uytq/oeen9uytq_flt.fits, ext 1: Scaling data by 0.8275521916478468 for temperature: 20.9769
./mastDownload/HST/oeen9velq/oeen9velq_flt.fits, ext 1: Scaling data by 0.928849220834831 for temperature: 19.0943
./mastDownload/HST/oeen9wf3q/oeen9wf3q_flt.fits, ext 1: Scaling data by 0.9506625167078937 for temperature: 18.7414
./mastDownload/HST/oeen9xjeq/oeen9xjeq_flt.fits, ext 1: Scaling data by 0.7139710570412897 for temperature: 23.7231
./mastDownload/HST/oeen9yjmq/oeen9yjmq_flt.fits, ext 1: Scaling data by 0.7139710570412897 for temperature: 23.7231
./mastDownload/HST/oeen9za2q/oeen9za2q_flt.fits, ext 1: Scaling data by 0.928849220834831 for temperature: 19.0943
./mastDownload/HST/oeena0aaq/oeena0aaq_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeena1g5q/oeena1g5q_flt.fits, ext 1: Scaling data by 0.9506625167078937 for temperature: 18.7414
./mastDownload/HST/oeena2gcq/oeena2gcq_flt.fits, ext 1: Scaling data by 0.9506625167078937 for temperature: 18.7414
./mastDownload/HST/oeena3kjq/oeena3kjq_flt.fits, ext 1: Scaling data by 0.9080202706445218 for temperature: 19.4471
./mastDownload/HST/oeena4knq/oeena4knq_flt.fits, ext 1: Scaling data by 0.8669560565983592 for temperature: 20.1923
Joining images
Performing CRREJECT
new_basedark_joined.fits
crcorr found = OMIT
FYI: CR rejection not already done
Keyword NRPTEXP = 1 while nr. of imsets = 56.0
>>>> Updated keyword NRPTEXP to 56.0
(and set keyword CRSPLIT to 1)
in new_basedark_joined.fits
Deleting old file: new_basedark_joined_bd_calstis_log.txt
Running CalSTIS on new_basedark_joined.fits
to create: new_basedark_joined_crj.fits
Normalizing by 61600.0
Cleaning...
basedark done for new_basedark.fits
with fits.open(new_basedark) as hdu:
new_basedark_data = hdu[1].data
cb = plt.imshow(new_basedark_data, cmap='plasma', vmax=1)
plt.colorbar(cb)
<matplotlib.colorbar.Colorbar at 0x7fc0ba07d750>
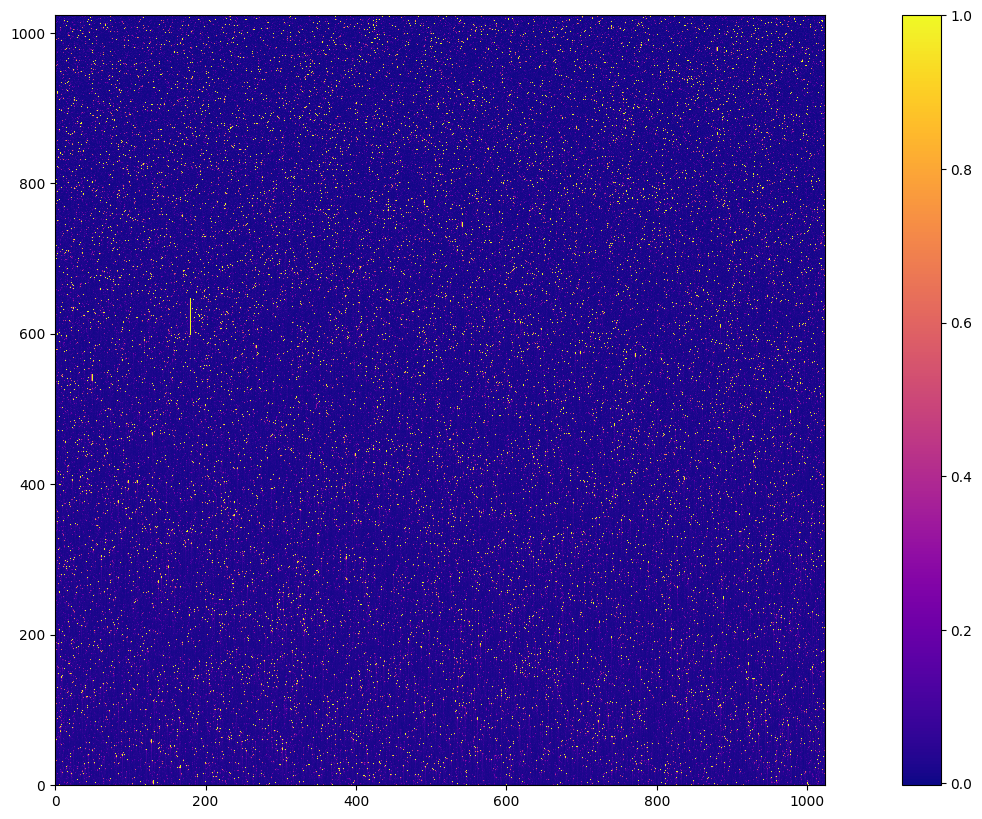
4. Make Weekdark#
The Weekdark is the combination of all “long” dark files from the week of the science observation, which is eventually passed into the Calstis pipline as the DARKFILE for the DARKCORR calibration. After the darks during a given week is combined and normalized to produce a weekly superdark, those hotpixels in the monthly Basedark are replaced by those of the normalized weekly superdark. The resulting dark has the high signal-to-noise ratio of the monthly baseline superdark, updated with the hot pixels of the current week.
We first search for the darks taken during the given week. We take the weekly period as the observation date ± 3 days in this demonstration, but notice here that since the observation time is 2021-05-05 12:29:54 while the ending time of the annealing period is 2021-05-05 14:00:22, observation date + 3 days will cross the annealing boundary. Therefore, in our case, we only take the observation date - 3 days as the weekly period. When you work with your own dataset, pay attention to the week boundary and annealing boundary to see if they completely overlap.
# search for darks taken during the weekly period (observation date, observation date + 3 days)
hdr = fits.getheader(crj, 0)
component_darks = Observations.query_criteria(
target_name='DARK',
t_min=[hdr['TEXPSTRT']-3, hdr['TEXPSTRT']],
t_exptime=[1099, 1101])
# get a list of files assiciated with that target
dark_list = Observations.get_product_list(component_darks)
dark_list = dark_list[dark_list['productSubGroupDescription'] == 'FLT']
# store all the paths to the superdark frames into a list
component_flt = [filename_mapping[x] for x in component_darks['obs_id']]
component_flt
[np.str_('./mastDownload/HST/oeen9za2q/oeen9za2q_flt.fits'),
np.str_('./mastDownload/HST/oeena0aaq/oeena0aaq_flt.fits'),
np.str_('./mastDownload/HST/oeena1g5q/oeena1g5q_flt.fits'),
np.str_('./mastDownload/HST/oeena2gcq/oeena2gcq_flt.fits'),
np.str_('./mastDownload/HST/oeena3kjq/oeena3kjq_flt.fits'),
np.str_('./mastDownload/HST/oeena4knq/oeena4knq_flt.fits')]
Now we have all the dark files we need to create the new weekdark reference file. Pass the list of the weekly component darks as the first parameter, the name of the new weekdark file as the second parameter, and the new basedark file we created above as the third parameter into refstis.make_weekdark. For more information on make_weekdark, see: Weekdark.
new_weekdark = "new_weekdark.fits"
# remove the new_basedark file if it already exists
if os.path.exists(new_weekdark):
os.remove(new_weekdark)
make_weekdark(component_flt, new_weekdark, thebasedark=new_basedark)
#-------------------------------#
# Running weekdark #
#-------------------------------#
Making weekdark new_weekdark.fits
With : oref$55a2044do_bia.fits
: new_basedark.fits
BIAS correction already done for ./mastDownload/HST/oeen9za2q/oeen9za2q_flt.fits
BIAS correction already done for ./mastDownload/HST/oeena0aaq/oeena0aaq_flt.fits
BIAS correction already done for ./mastDownload/HST/oeena1g5q/oeena1g5q_flt.fits
BIAS correction already done for ./mastDownload/HST/oeena2gcq/oeena2gcq_flt.fits
BIAS correction already done for ./mastDownload/HST/oeena3kjq/oeena3kjq_flt.fits
BIAS correction already done for ./mastDownload/HST/oeena4knq/oeena4knq_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeen9za2q/oeen9za2q_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeena0aaq/oeena0aaq_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeena1g5q/oeena1g5q_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeena2gcq/oeena2gcq_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeena3kjq/oeena3kjq_flt.fits
TEMPCORR = COMPLETE, no temperature correction applied to ./mastDownload/HST/oeena4knq/oeena4knq_flt.fits
Joining images to new_weekdark_joined.fits
new_weekdark_joined.fits
crcorr found = OMIT
FYI: CR rejection not already done
Keyword NRPTEXP = 1 while nr. of imsets = 6.0
>>>> Updated keyword NRPTEXP to 6.0
(and set keyword CRSPLIT to 1)
in new_weekdark_joined.fits
## crdone is 0
Deleting old file: new_weekdark_joined_bd_calstis_log.txt
Running CalSTIS on new_weekdark_joined.fits
to create: new_weekdark_joined_crj.fits
Normalizing by 6600.0
hot pixels are defined as above: 0.104120255
Cleaning up...
Weekdark done for new_weekdark.fits
5. Calibrate with New Weekdark#
5.1 Calibration#
Now we have created the new weekdark reference file for our specific dataset, we can use it to calibrate the raw data using Calstis. To change the dark reference file, we first set the value of DARKFILE in the _raw data 0th header using fits.setval. Calstis will then look for the DARKFILE value and use it as the reference file for DARKCORR.
raw = os.path.join("./mastDownload/HST", "{}".format(obs_id), "{}_raw.fits".format(obs_id))
wav = os.path.join("./mastDownload/HST", "{}".format(obs_id), "{}_wav.fits".format(obs_id))
# set the value of DARKFILE to the filename of the new week dark
fits.setval(raw, ext=0, keyword='DARKFILE', value=new_weekdark)
# make sure that the value is set correctly
fits.getval(raw, ext=0, keyword='DARKFILE')
'new_weekdark.fits'
Calibrate the _raw data using the new weekdark reference file:
%%capture --no-stderr output
# create a new folder to store the calibrated data
if os.path.exists('./new_dark'):
shutil.rmtree('./new_dark')
Path('./new_dark').mkdir(exist_ok=True)
res = stistools.calstis.calstis(raw, wavecal=wav, outroot="./new_dark/")
assert res == 0, 'CalSTIS returned an error!'
*** CALSTIS-0 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:22 UTC
Input ./mastDownload/HST/oeik1s030/oeik1s030_raw.fits
Outroot ./new_dark/oeik1s030_raw.fits
Wavecal ./mastDownload/HST/oeik1s030/oeik1s030_wav.fits
Warning DARKFILE is not the same in the science and wavecal headers; \
Warning values in the science and wavecal are respectively: \
Warning `new_weekdark.fits' \
Warning `oref$55a20445o_drk.fits' \
Warning The value from the science header will be used for the wavecal.
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:22 UTC
Input ./mastDownload/HST/oeik1s030/oeik1s030_raw.fits
Output ./new_dark/oeik1s030_blv_tmp.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:22 UTC
Epcfile oeik1snkj_epc.fits
Warning EPCTAB `oeik1snkj_epc.fits' not found.
CCDTAB oref$16j1600do_ccd.fits
CCDTAB PEDIGREE=GROUND
CCDTAB DESCRIP =Updated amp=D gain=4 atodgain and corresponding readnoise values---
CCDTAB DESCRIP =Oct. 1996 Air Calibration
DQICORR PERFORM
DQITAB oref$h1v11475o_bpx.fits
DQITAB PEDIGREE=GROUND
DQITAB DESCRIP =Prel. Ground Calib
DQICORR COMPLETE
ATODCORR OMIT
BLEVCORR PERFORM
Bias level from overscan has been subtracted; \
mean of bias levels subtracted was 1541.32.
BLEVCORR COMPLETE
Uncertainty array initialized, readnoise=8.2328, gain=4.016
BIASCORR PERFORM
BIASFILE oref$55a20443o_bia.fits
BIASFILE PEDIGREE=INFLIGHT 22/04/2021 05/05/2021
BIASFILE DESCRIP =Refbias (4/D) for data taken after 2021-04-21----------------------
BIASCORR COMPLETE
DARKCORR OMIT
FLATCORR OMIT
SHADCORR OMIT
PHOTCORR OMIT
Imset 1 End 20:17:22 UTC
Imset 2 Begin 20:17:22 UTC
Epcfile oeik1snlj_epc.fits
Warning EPCTAB `oeik1snlj_epc.fits' not found.
DQICORR PERFORM
DQICORR COMPLETE
ATODCORR OMIT
BLEVCORR PERFORM
Bias level from overscan has been subtracted; \
mean of bias levels subtracted was 1541.93.
BLEVCORR COMPLETE
Uncertainty array initialized, readnoise=8.2328, gain=4.016
BIASCORR PERFORM
BIASCORR COMPLETE
DARKCORR OMIT
FLATCORR OMIT
SHADCORR OMIT
Imset 2 End 20:17:22 UTC
Imset 3 Begin 20:17:22 UTC
Epcfile oeik1snmj_epc.fits
Warning EPCTAB `oeik1snmj_epc.fits' not found.
DQICORR PERFORM
DQICORR COMPLETE
ATODCORR OMIT
BLEVCORR PERFORM
Bias level from overscan has been subtracted; \
mean of bias levels subtracted was 1541.66.
BLEVCORR COMPLETE
Uncertainty array initialized, readnoise=8.2328, gain=4.016
BIASCORR PERFORM
BIASCORR COMPLETE
DARKCORR OMIT
FLATCORR OMIT
SHADCORR OMIT
Imset 3 End 20:17:22 UTC
Imset 4 Begin 20:17:22 UTC
Epcfile oeik1snnj_epc.fits
Warning EPCTAB `oeik1snnj_epc.fits' not found.
DQICORR PERFORM
DQICORR COMPLETE
ATODCORR OMIT
BLEVCORR PERFORM
Bias level from overscan has been subtracted; \
mean of bias levels subtracted was 1541.41.
BLEVCORR COMPLETE
Uncertainty array initialized, readnoise=8.2328, gain=4.016
BIASCORR PERFORM
BIASCORR COMPLETE
DARKCORR OMIT
FLATCORR OMIT
SHADCORR OMIT
Imset 4 End 20:17:22 UTC
End 02-Dec-2025 20:17:22 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-2 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:22 UTC
Input ./new_dark/oeik1s030_blv_tmp.fits
Output ./new_dark/oeik1s030_crj_tmp.fits
CRCORR PERFORM
Total number of input image sets = 4
CRREJTAB oref$j3m1403io_crr.fits
CRCORR COMPLETE
End 02-Dec-2025 20:17:22 UTC
*** CALSTIS-2 complete ***
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:22 UTC
Input ./new_dark/oeik1s030_crj_tmp.fits
Output ./new_dark/oeik1s030_crj.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:22 UTC
CCDTAB oref$16j1600do_ccd.fits
CCDTAB PEDIGREE=GROUND
CCDTAB DESCRIP =Updated amp=D gain=4 atodgain and corresponding readnoise values---
CCDTAB DESCRIP =Oct. 1996 Air Calibration
DQICORR OMIT
ATODCORR OMIT
BLEVCORR OMIT
BIASCORR OMIT
DARKCORR PERFORM
DARKFILE new_weekdark.fits
DARKFILE PEDIGREE=INFLIGHT 03/05/2021 05/05/2021
DARKFILE DESCRIP =Weekly gain=1 dark for STIS CCD data taken after May 03 2021-------
DARKCORR COMPLETE
FLATCORR PERFORM
PFLTFILE oref$x6417096o_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 24/10/2011 24/06/2012
PFLTFILE DESCRIP =REVISED ON-ORBIT STIS SPECTROSCOPIC CCD P-FLAT FOR L-MODES---------
LFLTFILE oref$pcc2026ko_lfl.fits
LFLTFILE PEDIGREE=INFLIGHT 27/05/1997 27/05/1997
LFLTFILE DESCRIP =CCD G750L Low-Order Flat
FLATCORR COMPLETE
SHADCORR OMIT
PHOTCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 1 End 20:17:23 UTC
End 02-Dec-2025 20:17:23 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:23 UTC
Input ./new_dark/oeik1s030_blv_tmp.fits
Output ./new_dark/oeik1s030_flt.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:23 UTC
Epcfile oeik1snkj_epc.fits
Warning EPCTAB `oeik1snkj_epc.fits' not found.
CCDTAB oref$16j1600do_ccd.fits
CCDTAB PEDIGREE=GROUND
CCDTAB DESCRIP =Updated amp=D gain=4 atodgain and corresponding readnoise values---
CCDTAB DESCRIP =Oct. 1996 Air Calibration
DQICORR OMIT
ATODCORR OMIT
BLEVCORR OMIT
BIASCORR OMIT
DARKCORR PERFORM
DARKFILE new_weekdark.fits
DARKFILE PEDIGREE=INFLIGHT 03/05/2021 05/05/2021
DARKFILE DESCRIP =Weekly gain=1 dark for STIS CCD data taken after May 03 2021-------
DARKCORR COMPLETE
FLATCORR PERFORM
PFLTFILE oref$x6417096o_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 24/10/2011 24/06/2012
PFLTFILE DESCRIP =REVISED ON-ORBIT STIS SPECTROSCOPIC CCD P-FLAT FOR L-MODES---------
LFLTFILE oref$pcc2026ko_lfl.fits
LFLTFILE PEDIGREE=INFLIGHT 27/05/1997 27/05/1997
LFLTFILE DESCRIP =CCD G750L Low-Order Flat
FLATCORR COMPLETE
SHADCORR OMIT
PHOTCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 1 End 20:17:23 UTC
Imset 2 Begin 20:17:23 UTC
Epcfile oeik1snlj_epc.fits
Warning EPCTAB `oeik1snlj_epc.fits' not found.
DQICORR OMIT
ATODCORR OMIT
BLEVCORR OMIT
BIASCORR OMIT
DARKCORR PERFORM
DARKCORR COMPLETE
FLATCORR PERFORM
FLATCORR COMPLETE
SHADCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 2 End 20:17:23 UTC
Imset 3 Begin 20:17:23 UTC
Epcfile oeik1snmj_epc.fits
Warning EPCTAB `oeik1snmj_epc.fits' not found.
DQICORR OMIT
ATODCORR OMIT
BLEVCORR OMIT
BIASCORR OMIT
DARKCORR PERFORM
DARKCORR COMPLETE
FLATCORR PERFORM
FLATCORR COMPLETE
SHADCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 3 End 20:17:23 UTC
Imset 4 Begin 20:17:23 UTC
Epcfile oeik1snnj_epc.fits
Warning EPCTAB `oeik1snnj_epc.fits' not found.
DQICORR OMIT
ATODCORR OMIT
BLEVCORR OMIT
BIASCORR OMIT
DARKCORR PERFORM
DARKCORR COMPLETE
FLATCORR PERFORM
FLATCORR COMPLETE
SHADCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 4 End 20:17:23 UTC
End 02-Dec-2025 20:17:23 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-1 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:23 UTC
Input ./mastDownload/HST/oeik1s030/oeik1s030_wav.fits
Output ./new_dark/oeik1s030_fwv_tmp.fits
OBSMODE ACCUM
APERTURE 52X0.1
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:23 UTC
Epcfile oeik1snoj_epc.fits
Warning EPCTAB `oeik1snoj_epc.fits' not found.
CCDTAB oref$16j1600do_ccd.fits
CCDTAB PEDIGREE=GROUND
CCDTAB DESCRIP =Updated amp=D gain=4 atodgain and corresponding readnoise values---
CCDTAB DESCRIP =Oct. 1996 Air Calibration
DQICORR PERFORM
DQITAB oref$h1v11475o_bpx.fits
DQITAB PEDIGREE=GROUND
DQITAB DESCRIP =Prel. Ground Calib
DQICORR COMPLETE
ATODCORR OMIT
BLEVCORR PERFORM
Bias level from overscan has been subtracted; \
mean of bias levels subtracted was 1540.52.
BLEVCORR COMPLETE
Uncertainty array initialized, readnoise=8.2328, gain=4.016
BIASCORR PERFORM
BIASFILE oref$55a20443o_bia.fits
BIASFILE PEDIGREE=INFLIGHT 22/04/2021 05/05/2021
BIASFILE DESCRIP =Refbias (4/D) for data taken after 2021-04-21----------------------
BIASCORR COMPLETE
DARKCORR PERFORM
DARKFILE new_weekdark.fits
DARKFILE PEDIGREE=INFLIGHT 03/05/2021 05/05/2021
DARKFILE DESCRIP =Weekly gain=1 dark for STIS CCD data taken after May 03 2021-------
DARKCORR COMPLETE
FLATCORR PERFORM
PFLTFILE oref$x6417096o_pfl.fits
PFLTFILE PEDIGREE=INFLIGHT 24/10/2011 24/06/2012
PFLTFILE DESCRIP =REVISED ON-ORBIT STIS SPECTROSCOPIC CCD P-FLAT FOR L-MODES---------
LFLTFILE oref$pcc2026ko_lfl.fits
LFLTFILE PEDIGREE=INFLIGHT 27/05/1997 27/05/1997
LFLTFILE DESCRIP =CCD G750L Low-Order Flat
FLATCORR COMPLETE
SHADCORR OMIT
PHOTCORR OMIT
STATFLAG PERFORM
STATFLAG COMPLETE
Imset 1 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-1 complete ***
*** CALSTIS-7 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Input ./new_dark/oeik1s030_fwv_tmp.fits
Output ./new_dark/oeik1s030_w2d_tmp.fits
OBSMODE ACCUM
APERTURE 52X0.1
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:24 UTC
Order 1 Begin 20:17:24 UTC
X2DCORR PERFORM
DISPCORR PERFORM
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
SDCTAB oref$16j16006o_sdc.fits
SDCTAB PEDIGREE=INFLIGHT 27/05/1997 13/06/2017
SDCTAB DESCRIP =Co-aligned fiducial bars via an update to the CDELT2 plate scales.-
SDCTAB DESCRIP =CDELT2 updated with inflight data, others Lindler/prelaunch
DISPTAB oref$l2j0137to_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 27/02/1997 24/11/1999
DISPTAB DESCRIP =Lindler, May 2000
DISPTAB DESCRIP =Lindler Postlaunch, May 2000
INANGTAB oref$h5s11397o_iac.fits
INANGTAB PEDIGREE=GROUND
INANGTAB DESCRIP =Model/C. Bowers May 6, 1997
INANGTAB DESCRIP =Model/C. Bowers
SPTRCTAB oref$qa31608go_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 13/02/1998
SPTRCTAB DESCRIP =New traces to account for angle change with time
SPTRCTAB DESCRIP =Lindler/Bohlin/Dressel/Holfeltz postlaunch calibration
X2DCORR COMPLETE
DISPCORR COMPLETE
Order 1 End 20:17:24 UTC
Imset 1 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-7 complete ***
*** CALSTIS-4 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Input ./new_dark/oeik1s030_w2d_tmp.fits
OBSMODE ACCUM
APERTURE 52X0.1
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:24 UTC
WAVECORR PERFORM
WCPTAB oref$16j1600co_wcp.fits
WCPTAB PEDIGREE=GROUND
WCPTAB DESCRIP =Force G140M and G230L SHIFTA2 into single-bar mode via SP_TRIM2.---
WCPTAB DESCRIP =TRIM values modified
LAMPTAB oref$l421050oo_lmp.fits
LAMPTAB PEDIGREE=GROUND
LAMPTAB DESCRIP =Ground+Inflight Cal PT lamp, 3.8 & 10 mA
LAMPTAB DESCRIP =Ground Cal for non-PRISM, PT lamp 10 mA
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
FLAGCR PERFORM
FLAGCR COMPLETE
Shift in dispersion direction is 2.957 pixels.
Shift in spatial direction is -1.950 pixels.
WAVECORR COMPLETE
Imset 1 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-4 complete ***
*** CALSTIS-12 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Wavecal ./new_dark/oeik1s030_w2d_tmp.fits
Science ./new_dark/oeik1s030_crj.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:24 UTC
SHIFTA1 set to 2.95707
SHIFTA2 set to -1.94995
Imset 1 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-12 complete ***
*** CALSTIS-12 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Wavecal ./new_dark/oeik1s030_w2d_tmp.fits
Science ./new_dark/oeik1s030_flt.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:24 UTC
SHIFTA1 set to 2.95707
SHIFTA2 set to -1.94995
Imset 1 End 20:17:24 UTC
Imset 2 Begin 20:17:24 UTC
SHIFTA1 set to 2.95707
SHIFTA2 set to -1.94995
Imset 2 End 20:17:24 UTC
Imset 3 Begin 20:17:24 UTC
SHIFTA1 set to 2.95707
SHIFTA2 set to -1.94995
Imset 3 End 20:17:24 UTC
Imset 4 Begin 20:17:24 UTC
SHIFTA1 set to 2.95707
SHIFTA2 set to -1.94995
Imset 4 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-12 complete ***
*** CALSTIS-7 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Input ./new_dark/oeik1s030_crj.fits
Output ./new_dark/oeik1s030_sx2.fits
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
Imset 1 Begin 20:17:24 UTC
HELCORR PERFORM
HELCORR COMPLETE
Order 1 Begin 20:17:24 UTC
X2DCORR PERFORM
DISPCORR PERFORM
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
SDCTAB oref$16j16006o_sdc.fits
SDCTAB PEDIGREE=INFLIGHT 27/05/1997 13/06/2017
SDCTAB DESCRIP =Co-aligned fiducial bars via an update to the CDELT2 plate scales.-
SDCTAB DESCRIP =CDELT2 updated with inflight data, others Lindler/prelaunch
DISPTAB oref$l2j0137to_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 27/02/1997 24/11/1999
DISPTAB DESCRIP =Lindler, May 2000
DISPTAB DESCRIP =Lindler Postlaunch, May 2000
INANGTAB oref$h5s11397o_iac.fits
INANGTAB PEDIGREE=GROUND
INANGTAB DESCRIP =Model/C. Bowers May 6, 1997
INANGTAB DESCRIP =Model/C. Bowers
SPTRCTAB oref$qa31608go_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 13/02/1998
SPTRCTAB DESCRIP =New traces to account for angle change with time
SPTRCTAB DESCRIP =Lindler/Bohlin/Dressel/Holfeltz postlaunch calibration
FLUXCORR PERFORM
PHOTTAB oref$95118575o_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 27/02/1997 14/04/1998
PHOTTAB DESCRIP =Fullerton 2025 calibration
PHOTTAB DESCRIP =Updated Sensitivities 2023
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$q541740po_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/5/1997 19/12/1998
PCTAB DESCRIP =Sensitivity for diff. extractions heights
PCTAB DESCRIP =12 STIS observations averaged
TDSTAB oref$8712049eo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/03/2024
TDSTAB DESCRIP =Updated time sensitivities for MIRVIS, G230LB, and G230MB
FLUXCORR COMPLETE
X2DCORR COMPLETE
DISPCORR COMPLETE
Order 1 End 20:17:24 UTC
Imset 1 End 20:17:24 UTC
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-7 complete ***
*** CALSTIS-6 -- Version 3.4.2 (19-Jan-2018) ***
Begin 02-Dec-2025 20:17:24 UTC
Input ./new_dark/oeik1s030_crj.fits
Output ./new_dark/oeik1s030_sx1.fits
Rootname oeik1s030
OBSMODE ACCUM
APERTURE 52X2
OPT_ELEM G750L
DETECTOR CCD
XTRACTAB oref$n7p1031qo_1dx.fits
XTRACTAB PEDIGREE=INFLIGHT 18/05/97
XTRACTAB DESCRIP =Analysis from prop. 7094
XTRACTAB DESCRIP =Analysis from prop. 7094
SPTRCTAB oref$qa31608go_1dt.fits
SPTRCTAB PEDIGREE=INFLIGHT 02/09/1997 13/02/1998
SPTRCTAB DESCRIP =New traces to account for angle change with time
Imset 1 Begin 20:17:24 UTC
Input read into memory.
Order 1 Begin 20:17:24 UTC
X1DCORR PERFORM
BACKCORR PERFORM
******** Calling Slfit ***********BACKCORR COMPLETE
X1DCORR COMPLETE
Spectrum extracted at y position = 510.673
DISPCORR PERFORM
DISPTAB oref$l2j0137to_dsp.fits
DISPTAB PEDIGREE=INFLIGHT 27/02/1997 24/11/1999
DISPTAB DESCRIP =Lindler, May 2000
DISPTAB DESCRIP =Lindler Postlaunch, May 2000
APDESTAB oref$16j16005o_apd.fits
APDESTAB PEDIGREE=INFLIGHT 01/03/1997 13/06/2017
APDESTAB DESCRIP =Aligned long-slit bar positions for single-bar cases.--------------
APDESTAB DESCRIP =Microscope Meas./Hartig Post-launch Offsets
INANGTAB oref$h5s11397o_iac.fits
INANGTAB DESCRIP =Model/C. Bowers May 6, 1997
DISPCORR COMPLETE
HELCORR PERFORM
HELCORR COMPLETE
PHOTTAB oref$95118575o_pht.fits
PHOTTAB PEDIGREE=INFLIGHT 27/02/1997 16/03/2021
PHOTTAB DESCRIP =Fullerton 2025 calibration
APERTAB oref$y2r1559to_apt.fits
APERTAB PEDIGREE=MODEL
APERTAB DESCRIP =Added/updated values for 31X0.05NDA,31X0.05NDB,31X0.05NDC apertures
APERTAB DESCRIP =Bohlin/Hartig TIM Models Nov. 1998
PCTAB oref$q541740po_pct.fits
PCTAB PEDIGREE=INFLIGHT 18/05/1997 19/12/1998
PCTAB DESCRIP =Sensitivity for diff. extractions heights
TDSTAB oref$8712049eo_tds.fits
TDSTAB PEDIGREE=INFLIGHT 01/04/1997 06/03/2024
TDSTAB DESCRIP =Updated time sensitivities for MIRVIS, G230LB, and G230MB
FLUXCORR PERFORM
CTECORR PERFORM
CCDTAB oref$16j1600do_ccd.fits
CCDTAB PEDIGREE=GROUND
CCDTAB DESCRIP =Updated amp=D gain=4 atodgain and corresponding readnoise values---
CCDTAB DESCRIP =Oct. 1996 Air Calibration
CTECORR COMPLETE
FLUXCORR COMPLETE
SGEOCORR OMIT
Row 1 written to disk.
Order 1 End 20:17:24 UTC
trace was rotated by = 0.0878885 degree.
Imset 1 End 20:17:24 UTC
Warning Keyword `XTRACALG' is being added to header.
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-6 complete ***
End 02-Dec-2025 20:17:24 UTC
*** CALSTIS-0 complete ***
with fits.open('new_weekdark.fits') as hdu:
new_weekdark_data = hdu[1].data
cb = plt.imshow(new_weekdark_data, cmap='plasma', vmax=1)
plt.colorbar(cb)
<matplotlib.colorbar.Colorbar at 0x7fc0ba09fb90>
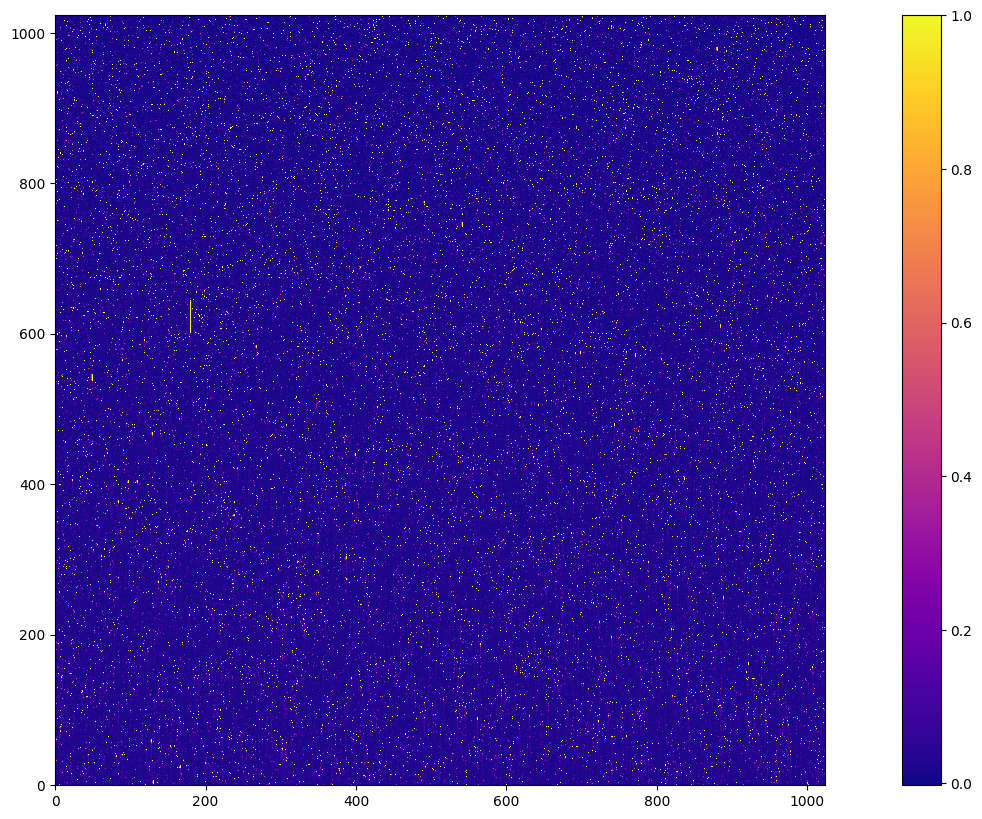
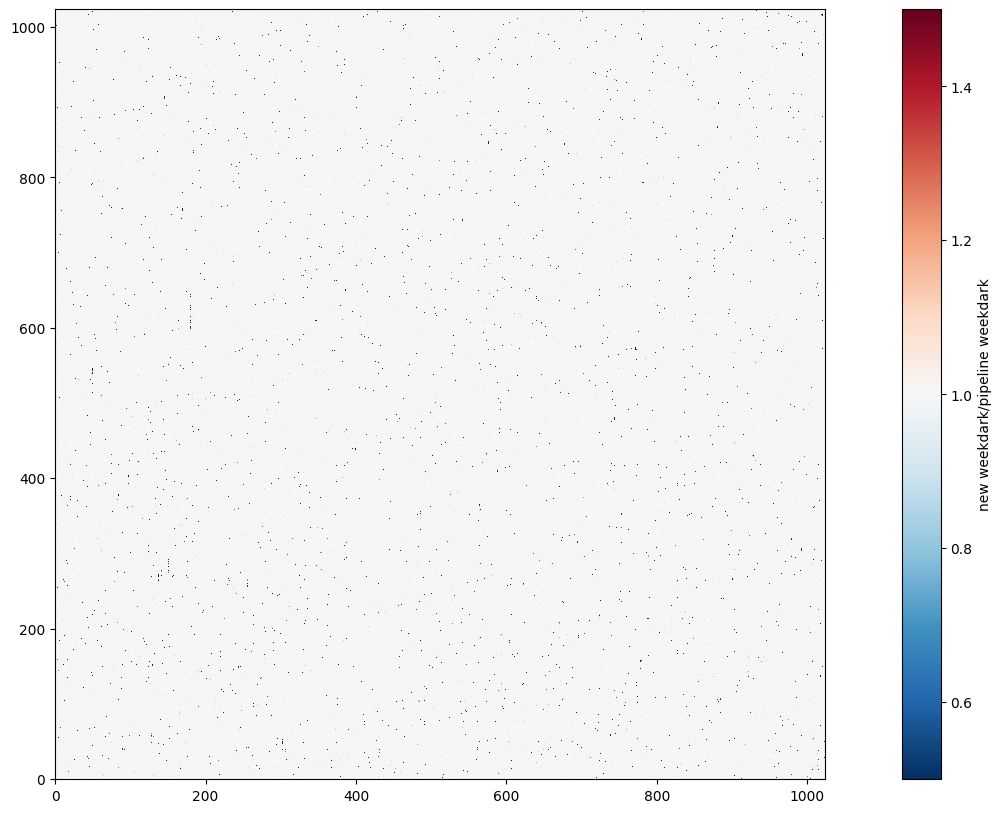
To compare the new weekdark science image with the old weekdark science image, we divide the new weekdark science image frame by that of the old weekdark, and use a diverging colormap to visulize the ratio. The colormap is normalized to center at 1, and the red pixels suggests that the ratio is greater than 1 while the blue pixels suggests that the ratio is less than 1. In general there are more blue pixels in the image, which means we are removing more hot pixels compared with the old weekdark.
with fits.open("55a20445o_drk.fits") as hdu:
old_weekdark_data = hdu[1].data
cb = plt.imshow(new_weekdark_data/old_weekdark_data, cmap='RdBu_r', vmin=0.5, vmax=1.5)
plt.colorbar(cb, label="new weekdark/pipeline weekdark")
<matplotlib.colorbar.Colorbar at 0x7fc0d23e45d0>
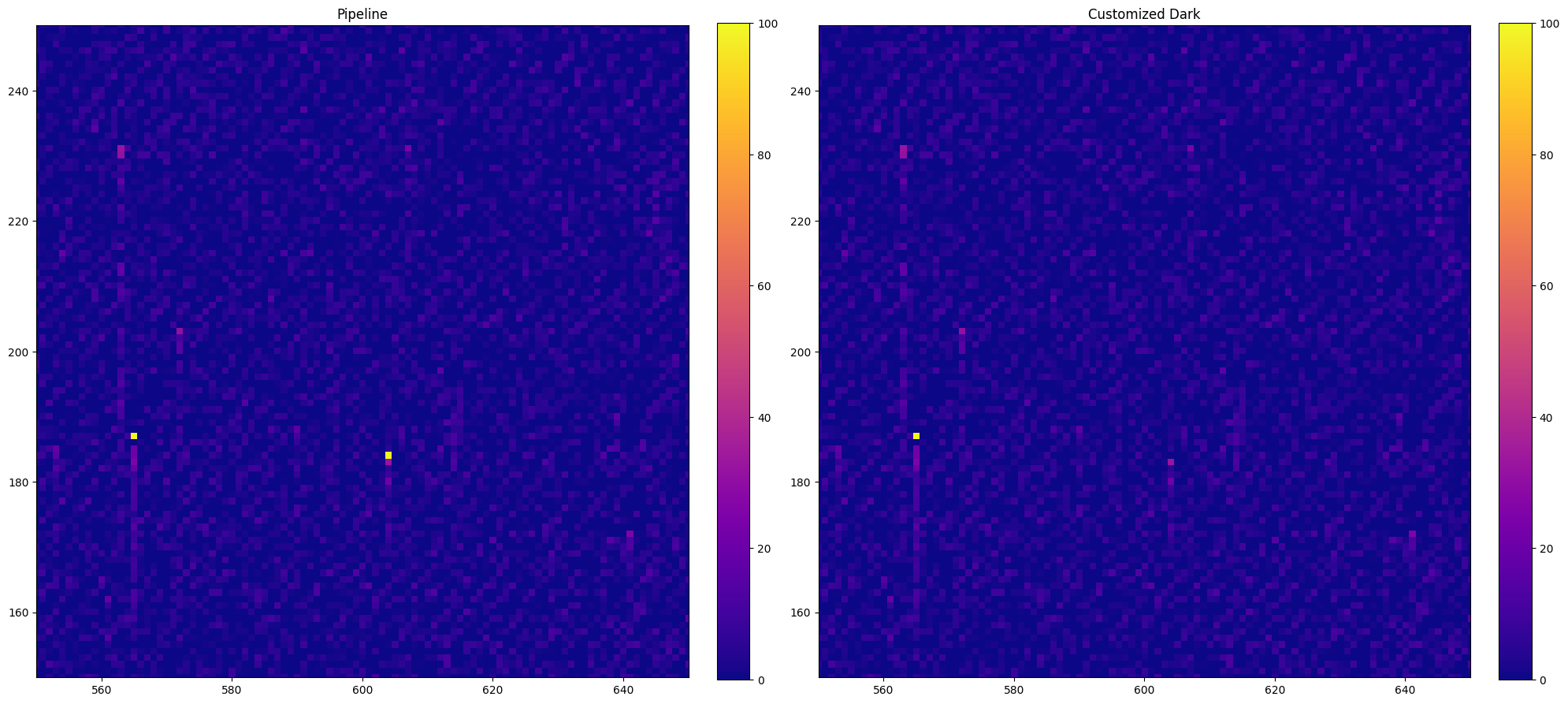
5.2 Comparison With the Default Dark File#
When we collected the science data from MAST, the _crj and _sx1 data files are already calibrated using the default dark reference file. We can make a comparison between the calibrated images and spectra of the defualt dark file and our new Weekdark. As shown in the comparison, a hot pixel is removed from the _crj image at x \(\approx\) 605 and y \(\approx\) 185.
# Plot the calibrated _crj images
# The left panel is the defalt _crj image from the pipline
# the right panel is calibrated with our customized dark file
plt.subplot(1, 2, 1)
with fits.open(crj) as hdu:
ex1 = hdu[1].data
cb = plt.imshow(ex1, vmin=0, vmax=100)
plt.colorbar(cb, fraction=0.046, pad=0.04)
plt.xlim(550, 650)
plt.ylim(150, 250)
plt.title("Pipeline")
plt.subplot(1, 2, 2)
with fits.open("./new_dark/oeik1s030_crj.fits") as hdu:
ex1 = hdu[1].data
cb = plt.imshow(ex1, vmin=0, vmax=100)
plt.colorbar(cb, fraction=0.046, pad=0.04)
plt.xlim(550, 650)
plt.ylim(150, 250)
plt.title("Customized Dark")
plt.tight_layout()
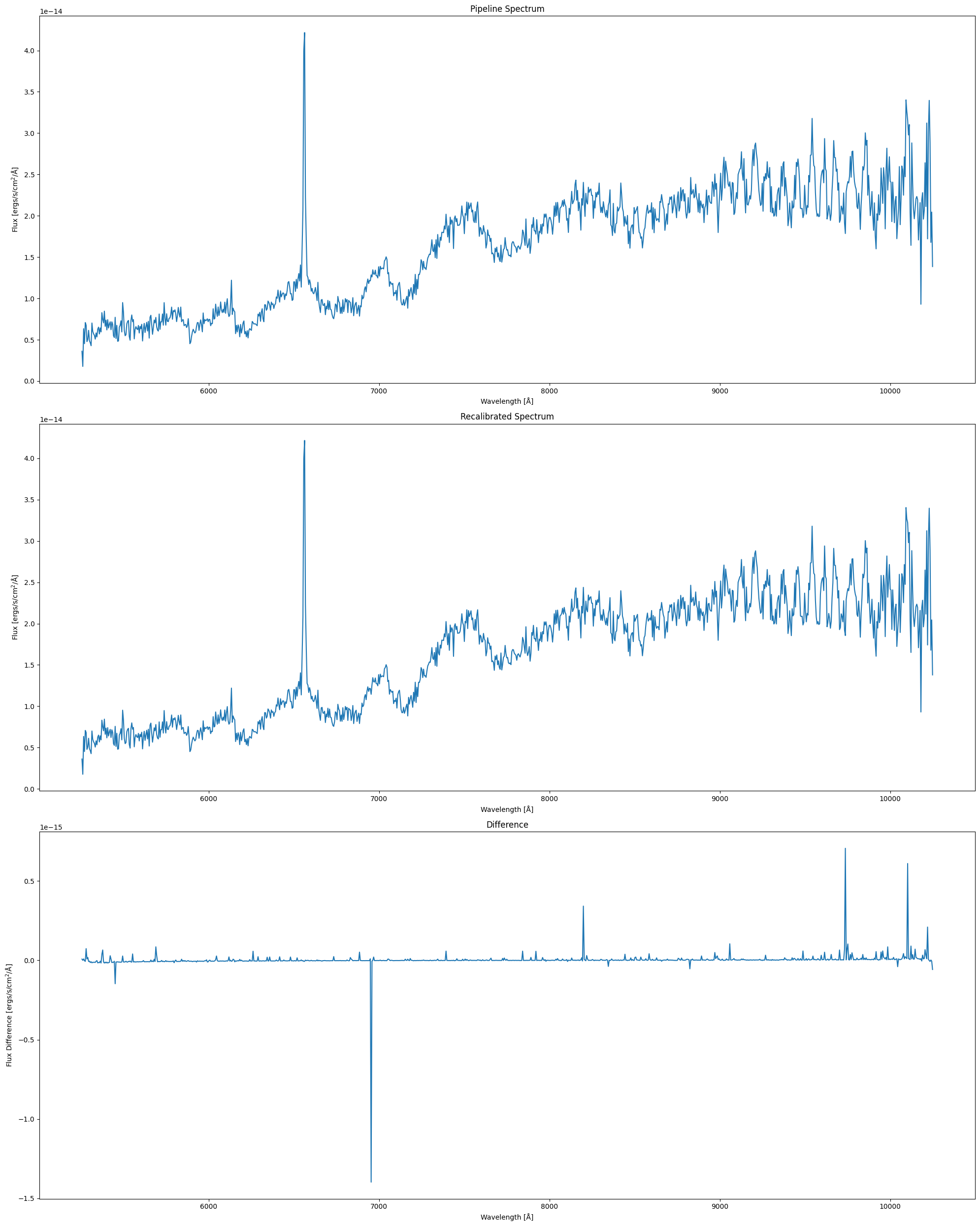
We can also visualize the flux difference in the _sx1 spectra in which we substrct the recalibrated spectrum by the pipeline spectrum:
plt.figure(figsize=(20, 25))
# get the spectrum of the default pipline _sx1 data
pip = Table.read("./mastDownload/HST/oeik1s030/oeik1s030_sx1.fits", 1)
wl, pip_flux = pip[0]["WAVELENGTH", "FLUX"]
# get the flux of the customized new_dark _sx1 data
cus = Table.read("./new_dark/oeik1s030_sx1.fits", 1)
cus_wl, cus_flux = cus[0]["WAVELENGTH", "FLUX"]
# interpolant flux so that the wavelengths matches
interp_flux = np.interp(wl, cus_wl, cus_flux)
# plot the pipeline spectrum
plt.subplot(3, 1, 1)
plt.plot(wl, pip_flux)
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Pipeline Spectrum")
# plot the pipeline spectrum
plt.subplot(3, 1, 2)
plt.plot(cus_wl, cus_flux)
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux [ergs/s/cm$^2$/Å]")
plt.title("Recalibrated Spectrum")
# plot the spectra difference
plt.subplot(3, 1, 3)
plt.plot(wl, interp_flux-pip_flux)
plt.xlabel("Wavelength [Å]")
plt.ylabel("Flux Difference [ergs/s/cm$^2$/Å]")
plt.title("Difference")
plt.tight_layout()
WARNING: UnitsWarning: 'Angstroms' did not parse as fits unit: At col 0, Unit 'Angstroms' not supported by the FITS standard. Did you mean Angstrom or angstrom? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Angstroms' did not parse as fits unit: At col 0, Unit 'Angstroms' not supported by the FITS standard. Did you mean Angstrom or angstrom? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
WARNING: UnitsWarning: 'Counts/s' did not parse as fits unit: At col 0, Unit 'Counts' not supported by the FITS standard. Did you mean count? If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html [astropy.units.core]
About this Notebook #
Author: Keyi Ding
Updated On: 2025-11-05
This tutorial was generated to be in compliance with the STScI style guides and would like to cite the Jupyter guide in particular.
Citations #
If you use astropy, matplotlib, astroquery, or numpy for published research, please cite the
authors. Follow these links for more information about citations:


