
WFC3/UVIS Time-dependent Photometry#
Learning Goals#
By the end of this tutorial, you will:
Compute aperture photometry on FLC frames acquired at three unique epochs and apply the new time-dependent inverse sensitivity (PHOTFLAM) keywords.
Recompute aperture photometry on modified FLC frames with ‘equalized’ countrate values.
Redrizzle the ‘equalized’ FLC frames and compute aperture photometry on the DRC products.
Table of Contents#
Introduction
1. Imports
2. Download the data
3. Correct for distortion using the Pixel Area Map
4. Compute aperture photometry on the FLC frames
5. Correct the FLC frames using ‘photometric equalization’
6. Redrizzle the corrected FLC frames for each epoch
7. Conclusions
Additional Resources
About the Notebook
Citations
Introduction#
For UVIS images retrieved after October 15, 2020, new time-dependent photometry keyword values (PHOTFLAM, PHTFLAM1, PHTFLAM2 and PHTRATIO) are populated in the image header and must be applied separately for each observation epoch. This is a change from prior calibration, where a single set of keyword values were provided for each filter, independent of date. For more detail on the new calibration, see WFC3 ISR 2021-04.
In this tutorial, we show how to use the time-dependent calibration to compute aperture photometry on UVIS calibrated, CTE-corrected images (flc.fits, hereafter FLC) obtained at three epochs, spanning a total range of ~8 years and showing a loss in sensitivity of ~2%. The repository includes a CSV file containing a list of FLCs and the centroid of the star in each image, as well as the UVIS Pixel Area Maps to correct for distortion when working with FLC data.
Alternately, the FLC science arrays may be ‘equalized’ to account for sensitivity changes prior to computing photometry, where a reference set of keywords may be then used for all images. This photometric ‘equalization’ must be performed before combining any set of FLC images with AstroDrizzle which span multiple epochs in time.
1. Imports#
This notebook assumes you have created the virtual environment in WFC3 Library’s installation instructions.
We import:
os for setting environment variables
glob for finding lists of files
shutil for managing directories
numpy for handling array functions
pandas for managing data
matplotlib.pyplot for plotting data
astroquery.mast Observations for downloading data from MAST
astropy for astronomy related functions
drizzlepac for combining images
photutils for photometric calculations
stwcs for updating the World Coordinate System
import os
import glob
import shutil
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.stats import sigma_clipped_stats
from astroquery.mast import Observations
from drizzlepac import photeq
from drizzlepac import astrodrizzle
from photutils.aperture import aperture_photometry, CircularAperture, CircularAnnulus
%matplotlib inline
2. Download the Data#
The following commands query MAST for WFC3/UVIS calibrated (FLC) data products in the F606W filter for three epochs of GD153 observations (acquired in 2009, 2013, and 2017) and then downloads them to the current directory.
data_list = Observations.query_criteria(obs_id=['IBCDA4010', 'ICH3040F0', 'IDBHA6040'])
Observations.download_products(data_list['obsid'], project='CALWF3',
mrp_only=False, download_dir='./data', productSubGroupDescription=['FLC', 'ASN'])
science_files = glob.glob('data/mastDownload/HST/*/*fits')
for im in science_files:
root = os.path.basename(im)
new_path = os.path.join("./", root)
os.rename(im, new_path)
shutil.rmtree('data/')
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibcda4d0q_flc.fits to ./data/mastDownload/HST/ibcda4d0q/ibcda4d0q_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibcda4d6q_flc.fits to ./data/mastDownload/HST/ibcda4d6q/ibcda4d6q_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibcda4dcq_flc.fits to ./data/mastDownload/HST/ibcda4dcq/ibcda4dcq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibcda4diq_flc.fits to ./data/mastDownload/HST/ibcda4diq/ibcda4diq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ibcda4010_asn.fits to ./data/mastDownload/HST/ibcda4010/ibcda4010_asn.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ich304prq_flc.fits to ./data/mastDownload/HST/ich304prq/ich304prq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ich304q1q_flc.fits to ./data/mastDownload/HST/ich304q1q/ich304q1q_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/ich3040f0_asn.fits to ./data/mastDownload/HST/ich3040f0/ich3040f0_asn.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/idbha6ntq_flc.fits to ./data/mastDownload/HST/idbha6ntq/idbha6ntq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/idbha6nyq_flc.fits to ./data/mastDownload/HST/idbha6nyq/idbha6nyq_flc.fits ...
[Done]
Downloading URL https://mast.stsci.edu/api/v0.1/Download/file?uri=mast:HST/product/idbha6040_asn.fits to ./data/mastDownload/HST/idbha6040/idbha6040_asn.fits ...
[Done]
For convenience, the file ‘GD153_F606W_public.csv’ contains information about the images required for the tutorial. For each FLC image, the following is provided: filename, star x-cdt, y-cdt, filter, CCD amplifier, chip, and epoch for the 3 observation dates.
df = pd.read_csv('GD153_F606W_public.csv')
df
| FLC | Centx | Centy | Filter | Amp | Chip | Epoch | |
|---|---|---|---|---|---|---|---|
| 0 | ibcda4d0q_flc.fits | 289.940 | 285.934 | F606W | C | 2 | 1 |
| 1 | ibcda4d6q_flc.fits | 325.855 | 299.742 | F606W | C | 2 | 1 |
| 2 | ibcda4dcq_flc.fits | 312.830 | 321.911 | F606W | C | 2 | 1 |
| 3 | ibcda4diq_flc.fits | 276.756 | 308.288 | F606W | C | 2 | 1 |
| 4 | ich304prq_flc.fits | 246.181 | 331.752 | F606W | C | 2 | 2 |
| 5 | ich304q1q_flc.fits | 248.901 | 334.022 | F606W | C | 2 | 2 |
| 6 | idbha6ntq_flc.fits | 262.719 | 254.764 | F606W | C | 2 | 3 |
| 7 | idbha6nyq_flc.fits | 265.020 | 257.049 | F606W | C | 2 | 3 |
3. Correct for distortion using the Pixel Area Map#
FLC frames are not corrected for distortion and pixels therefore do not have equal area on the sky. To correct for this affect, we multiply the FLC frames by the Pixel Area Map (PAM). Since the GD153 data are C512C subarrays, the PAM needs to be “cut out” at the region corresponding to the subarray.
pam1 = fits.getdata('UVIS1wfc3_map.fits')
pam2 = fits.getdata('UVIS2wfc3_map.fits')
pams = {'A': pam1[-512:, :513], 'B': pam1[-512:, -513:], 'C': pam2[:512, :513], 'D': pam2[:512, -513:]}
4. Compute aperture photometry on the FLC frames#
4.1 Calculate countrates#
Here we set up the code that computes the photometry. We are using a standard aperture size of 10 pixels, with a sky annulus from 155 to 165 pixels. These are the steps involved in computing the photometry:
Loop through each FLC and load the data, MJD date of exposure, exposure time, and PHOTFLAM.
Divide each FLC by the exposure time to get countrates in electrons per second (FLC images are in electrons).
Correct for distortion by multiplying by the appropriate cutout of the pixel area map (PAM) corresponding to the subarray region.
Supply the x,y coordinate of the star in each image from the CSV file/pandas dataframe.
Define the aperture, annulus aperture, and annulus mask.
Compute the sigma-clipped mean of the sky annulus.
Compute the photometry in the aperture.
Subtract the sky background from the photometry derived in the previous step.
Store all values.
phots = []
mjds = []
dates = []
ap = 10
skyrad = [155, 165]
pfl = []
for i, flc in enumerate(df['FLC'].values):
with fits.open(flc) as f:
data = f[1].data
mjd = f[0].header['EXPSTART']
date = f[0].header['DATE-OBS']
exptime = f[0].header['EXPTIME']
pfl.append(f[0].header['PHOTFLAM'])
data = data / exptime
data = data * pams[df.at[i, 'Amp']]
positions = (df.at[i, 'Centx'], df.at[i, 'Centy'])
aperture = CircularAperture(positions, ap)
annulus_aperture = CircularAnnulus(positions, r_in=skyrad[0], r_out=skyrad[1])
annulus_masks = annulus_aperture.to_mask(method='center')
annulus_data = annulus_masks.multiply(data)
mask = annulus_masks.data
annulus_data_1d = annulus_data[mask > 0]
mean_sigclip, _, _ = sigma_clipped_stats(annulus_data_1d)
apers = [aperture, annulus_aperture]
phot_table = aperture_photometry(data, apers)
background = mean_sigclip * aperture.area
final_sum = phot_table['aperture_sum_0'] - background
phots.append(final_sum[0])
mjds.append(mjd)
dates.append(date)
Next, we append the coutrates, MJD’s, and PHOTFLAM values to our dataframe.
df['Countrate'] = phots
df['MJD'] = mjds
df['DATE-OBS'] = dates
df['PHOTFLAM'] = pfl
df
| FLC | Centx | Centy | Filter | Amp | Chip | Epoch | Countrate | MJD | DATE-OBS | PHOTFLAM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ibcda4d0q_flc.fits | 289.940 | 285.934 | F606W | C | 2 | 1 | 103300.375759 | 55170.563332 | 2009-12-05 | 1.153991e-19 |
| 1 | ibcda4d6q_flc.fits | 325.855 | 299.742 | F606W | C | 2 | 1 | 103292.263099 | 55170.569026 | 2009-12-05 | 1.153991e-19 |
| 2 | ibcda4dcq_flc.fits | 312.830 | 321.911 | F606W | C | 2 | 1 | 103661.304094 | 55170.574721 | 2009-12-05 | 1.153991e-19 |
| 3 | ibcda4diq_flc.fits | 276.756 | 308.288 | F606W | C | 2 | 1 | 103448.275067 | 55170.620196 | 2009-12-05 | 1.153992e-19 |
| 4 | ich304prq_flc.fits | 246.181 | 331.752 | F606W | C | 2 | 2 | 102471.856752 | 56629.866744 | 2013-12-03 | 1.163887e-19 |
| 5 | ich304q1q_flc.fits | 248.901 | 334.022 | F606W | C | 2 | 2 | 102744.163768 | 56629.877033 | 2013-12-03 | 1.163887e-19 |
| 6 | idbha6ntq_flc.fits | 262.719 | 254.764 | F606W | C | 2 | 3 | 101822.822242 | 58068.856082 | 2017-11-11 | 1.173665e-19 |
| 7 | idbha6nyq_flc.fits | 265.020 | 257.049 | F606W | C | 2 | 3 | 101553.087031 | 58068.861359 | 2017-11-11 | 1.173665e-19 |
4.2 Calculate magnitudes#
We now convert countrates into ST magnitudes using the PHOTFLAM value and the following equation. The \(EE_{r10}\) is the encircled energy term for an aperture radius of r=10 pixels (0.4 arcseconds). For F606W with UVIS2, this is 0.91. This value can be computed using stsynphot as described in the ‘Photometry Examples’ notebook in this WFC3 Library repository.
EE_r10 = 0.91
df["STMags"] = (
- 21.1
- 2.5 * np.log10(df["PHOTFLAM"])
- 2.5 * np.log10(df["Countrate"])
- 2.5 * np.log10(1.0 / EE_r10)
)
df
| FLC | Centx | Centy | Filter | Amp | Chip | Epoch | Countrate | MJD | DATE-OBS | PHOTFLAM | STMags | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ibcda4d0q_flc.fits | 289.940 | 285.934 | F606W | C | 2 | 1 | 103300.375759 | 55170.563332 | 2009-12-05 | 1.153991e-19 | 13.606843 |
| 1 | ibcda4d6q_flc.fits | 325.855 | 299.742 | F606W | C | 2 | 1 | 103292.263099 | 55170.569026 | 2009-12-05 | 1.153991e-19 | 13.606928 |
| 2 | ibcda4dcq_flc.fits | 312.830 | 321.911 | F606W | C | 2 | 1 | 103661.304094 | 55170.574721 | 2009-12-05 | 1.153991e-19 | 13.603055 |
| 3 | ibcda4diq_flc.fits | 276.756 | 308.288 | F606W | C | 2 | 1 | 103448.275067 | 55170.620196 | 2009-12-05 | 1.153992e-19 | 13.605289 |
| 4 | ich304prq_flc.fits | 246.181 | 331.752 | F606W | C | 2 | 2 | 102471.856752 | 56629.866744 | 2013-12-03 | 1.163887e-19 | 13.606315 |
| 5 | ich304q1q_flc.fits | 248.901 | 334.022 | F606W | C | 2 | 2 | 102744.163768 | 56629.877033 | 2013-12-03 | 1.163887e-19 | 13.603434 |
| 6 | idbha6ntq_flc.fits | 262.719 | 254.764 | F606W | C | 2 | 3 | 101822.822242 | 58068.856082 | 2017-11-11 | 1.173665e-19 | 13.604131 |
| 7 | idbha6nyq_flc.fits | 265.020 | 257.049 | F606W | C | 2 | 3 | 101553.087031 | 58068.861359 | 2017-11-11 | 1.173665e-19 | 13.607010 |
4.3 Plot countrate vs date#
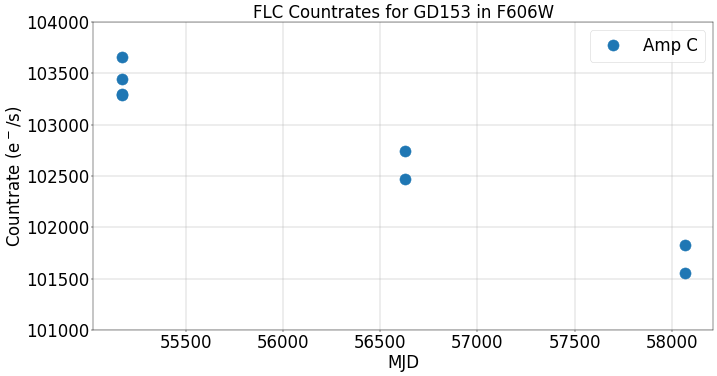
We first plot the photometric countrates (electrons per second) vs time in MJD. The decline in the observed countrate is due to sensitivity loss in F606W at a rate of ~0.2% per year.
fig = plt.figure(figsize=(20, 10), dpi=40)
plt.plot(df['MJD'], df['Countrate'], 'o', markersize=20, label='Amp C')
plt.grid()
plt.xlabel('MJD', fontsize=30)
plt.xticks(fontsize=30)
plt.yticks(fontsize=30)
plt.ylabel(r'Countrate (e$^-$/s)', fontsize=30)
plt.title('FLC Countrates for GD153 in F606W', fontsize=30)
plt.ylim(101000, 104000)
plt.legend(loc=0, fontsize=30)
<matplotlib.legend.Legend at 0x7fe5ed4056d0>
4.4 Plot magnitude vs date#
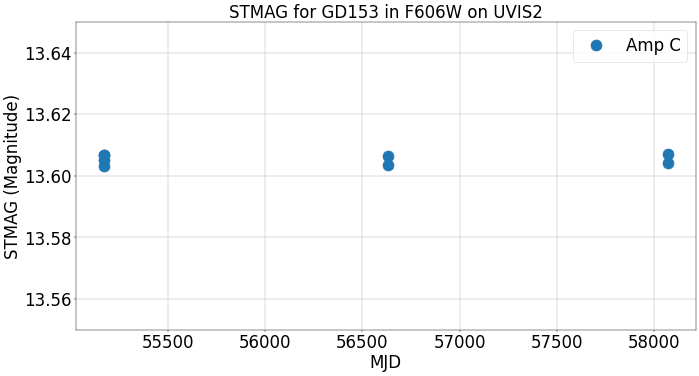
Now, we plot the ST magnitude versus time in MJD. This is computed using the ‘corrected’ PHOTFLAM keyword, so the magnitude values are stable over time.
fig = plt.figure(figsize=(20, 10), dpi=40)
plt.plot(df['MJD'], df['STMags'], 'o', markersize=20, label='Amp C')
plt.grid()
plt.ticklabel_format(useOffset=False)
plt.xlabel('MJD', fontsize=30)
plt.xticks(fontsize=30)
plt.yticks(fontsize=30)
plt.ylabel('STMAG (Magnitude)', fontsize=30)
plt.ylim(13.55, 13.65)
plt.title('STMAG for GD153 in F606W on UVIS2', fontsize=30)
plt.legend(loc=0, fontsize=30)
<matplotlib.legend.Legend at 0x7fe5eb108490>
5. Correct the FLC frames using ‘photometric equalization’#
This single-line step will equalize the countrates in the science array of the FLC frames to match any specified ‘reference’ image. For more details, see the drizzlepac documentation for photeq. Note that at this step, we overwrite the science pixels in the original FLC files. In this case the data are sorted, and the software automatically uses the PHOTFLAM value of the first 2009 image as a reference for matching the with the other images. You can supply a given reference PHOTFLAM value to the photeq call and/or ensure that your data are time-sorted.
Note: Running this cell will edit the FLCs in the local directory, and you will need to download the files again if you require the original data.
photeq.photeq(','.join(df['FLC'].values), readonly=False)
***** photeq started on 2025-12-02 20:16:01.246717
Version 3.10.0
PRIMARY PHOTOMETRIC KEYWORD: PHOTFLAM
SECONDARY PHOTOMETRIC KEYWORD(S): PHOTFNU
REFERENCE VALUE FROM FILE: 'ibcda4d0q_flc.fits[0]'
REFERENCE 'PHOTFLAM' VALUE IS: 1.1539911e-19
Processing file 'ibcda4d0q_flc.fits'
* Primary header:
- 'PHOTFLAM' = 1.1539911e-19 found in the primary header.
- Data conversion factor based on primary header: 1.0
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1539911e-19)
- Computed conversion factor for data: 1.0
- Setting PHOTFNU to 1.3456969e-07 (old value was 1.3456969e-07)
- Data have been multiplied by 1.0
- Error array (ext=('ERR', 1)) has been multiplied by 1.0
Processing file 'ibcda4d6q_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1539913e-19)
- Data conversion factor based on primary header: 1.0000001733115618
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1539913e-19)
- Computed conversion factor for data: 1.0000001733115618
- Setting PHOTFNU to 1.345696666775209e-07 (old value was 1.3456969e-07)
- Data have been multiplied by 1.0000001733115618
- Error array (ext=('ERR', 1)) has been multiplied by 1.0000001733115618
Processing file 'ibcda4dcq_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1539913e-19)
- Data conversion factor based on primary header: 1.0000001733115618
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1539913e-19)
- Computed conversion factor for data: 1.0000001733115618
- Setting PHOTFNU to 1.345696666775209e-07 (old value was 1.3456969e-07)
- Data have been multiplied by 1.0000001733115618
- Error array (ext=('ERR', 1)) has been multiplied by 1.0000001733115618
Processing file 'ibcda4diq_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1539915e-19)
- Data conversion factor based on primary header: 1.000000346623124
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1539915e-19)
- Computed conversion factor for data: 1.000000346623124
- Setting PHOTFNU to 1.3456968335503596e-07 (old value was 1.3456973e-07)
- Data have been multiplied by 1.000000346623124
- Error array (ext=('ERR', 1)) has been multiplied by 1.000000346623124
Processing file 'ich304prq_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1638865e-19)
- Data conversion factor based on primary header: 1.0085749361498544
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1638865e-19)
- Computed conversion factor for data: 1.0085749361498544
- Setting PHOTFNU to 1.3434231324172846e-07 (old value was 1.3549429e-07)
- Data have been multiplied by 1.0085749361498544
- Error array (ext=('ERR', 1)) has been multiplied by 1.0085749361498544
Processing file 'ich304q1q_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1638865e-19)
- Data conversion factor based on primary header: 1.0085749361498544
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1638865e-19)
- Computed conversion factor for data: 1.0085749361498544
- Setting PHOTFNU to 1.3434232315670814e-07 (old value was 1.354943e-07)
- Data have been multiplied by 1.0085749361498544
- Error array (ext=('ERR', 1)) has been multiplied by 1.0085749361498544
Processing file 'idbha6ntq_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1736647e-19)
- Data conversion factor based on primary header: 1.0170483117244147
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1736647e-19)
- Computed conversion factor for data: 1.0170483117244147
- Setting PHOTFNU to 1.3411951863793208e-07 (old value was 1.3640603e-07)
- Data have been multiplied by 1.0170483117244147
- Error array (ext=('ERR', 1)) has been multiplied by 1.0170483117244147
Processing file 'idbha6nyq_flc.fits'
* Primary header:
- 'PHOTFLAM' found in the primary header.
- Setting PHOTFLAM in the primary header to 1.1539911e-19 (old value was 1.1736649e-19)
- Data conversion factor based on primary header: 1.0170484850359764
* EXT: ('SCI', 1)
- Setting PHOTFLAM to 1.1539911e-19 (old value was 1.1736649e-19)
- Computed conversion factor for data: 1.0170484850359764
- Setting PHOTFNU to 1.3411949578310894e-07 (old value was 1.3640603e-07)
- Data have been multiplied by 1.0170484850359764
- Error array (ext=('ERR', 1)) has been multiplied by 1.0170484850359764
Done.
5.1 Recompute countrates#
We repeat the same aperture photometry as in Section 4.1 but using the photometrically equalized FLC data.
phots = []
for i, flc in enumerate(df['FLC'].values):
with fits.open(flc) as f:
data = f[1].data
exptime = f[0].header['EXPTIME']
data = data / exptime
data = data * pams[df.at[i, 'Amp']]
positions = (df.at[i, 'Centx'], df.at[i, 'Centy'])
aperture = CircularAperture(positions, ap)
annulus_aperture = CircularAnnulus(positions, r_in=skyrad[0], r_out=skyrad[1])
annulus_masks = annulus_aperture.to_mask(method='center')
annulus_data = annulus_masks.multiply(data)
mask = annulus_masks.data
annulus_data_1d = annulus_data[mask > 0]
mean_sigclip, _, _ = sigma_clipped_stats(annulus_data_1d)
background = mean_sigclip * aperture.area
apers = [aperture, annulus_aperture]
phot_table = aperture_photometry(data, apers)
final_sum = phot_table['aperture_sum_0'] - background
phots.append(final_sum[0])
mjds.append(mjd)
We’ll append the photometrically equalized countrates to our dataframe.
df['Phot-eq'] = phots
df
| FLC | Centx | Centy | Filter | Amp | Chip | Epoch | Countrate | MJD | DATE-OBS | PHOTFLAM | STMags | Phot-eq | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ibcda4d0q_flc.fits | 289.940 | 285.934 | F606W | C | 2 | 1 | 103300.375759 | 55170.563332 | 2009-12-05 | 1.153991e-19 | 13.606843 | 103300.375759 |
| 1 | ibcda4d6q_flc.fits | 325.855 | 299.742 | F606W | C | 2 | 1 | 103292.263099 | 55170.569026 | 2009-12-05 | 1.153991e-19 | 13.606928 | 103292.274260 |
| 2 | ibcda4dcq_flc.fits | 312.830 | 321.911 | F606W | C | 2 | 1 | 103661.304094 | 55170.574721 | 2009-12-05 | 1.153991e-19 | 13.603055 | 103661.316241 |
| 3 | ibcda4diq_flc.fits | 276.756 | 308.288 | F606W | C | 2 | 1 | 103448.275067 | 55170.620196 | 2009-12-05 | 1.153992e-19 | 13.605289 | 103448.313094 |
| 4 | ich304prq_flc.fits | 246.181 | 331.752 | F606W | C | 2 | 2 | 102471.856752 | 56629.866744 | 2013-12-03 | 1.163887e-19 | 13.606315 | 103350.548848 |
| 5 | ich304q1q_flc.fits | 248.901 | 334.022 | F606W | C | 2 | 2 | 102744.163768 | 56629.877033 | 2013-12-03 | 1.163887e-19 | 13.603434 | 103625.191460 |
| 6 | idbha6ntq_flc.fits | 262.719 | 254.764 | F606W | C | 2 | 3 | 101822.822242 | 58068.856082 | 2017-11-11 | 1.173665e-19 | 13.604131 | 103558.735918 |
| 7 | idbha6nyq_flc.fits | 265.020 | 257.049 | F606W | C | 2 | 3 | 101553.087031 | 58068.861359 | 2017-11-11 | 1.173665e-19 | 13.607010 | 103284.414246 |
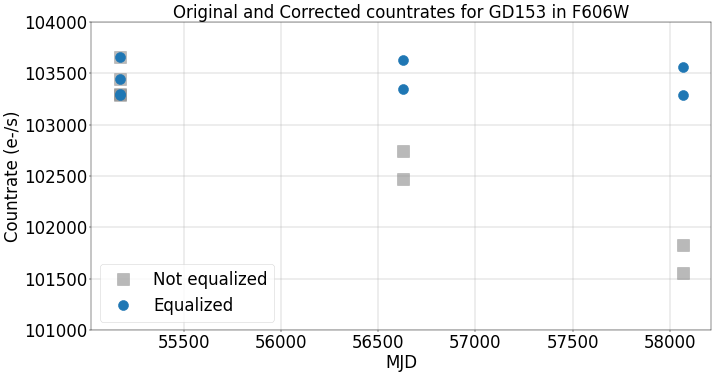
5.2 Plot corrected countrate vs date#
We plot the photometric countrate (electrons per second) versus time in MJD. The corrected data in blue shows that the countrate is now ~flat over time to within the measurment errors. The original countrates are shown in grey and show a decline of nearly 2% over the three epochs.
fig = plt.figure(figsize=(20, 10), dpi=40)
plt.plot(df['MJD'], df['Countrate'], 's', markersize=20, label='Not equalized', alpha=0.55, color='Grey')
plt.plot(df['MJD'], df['Phot-eq'], 'o', markersize=18, label='Equalized')
plt.grid()
plt.xlabel('MJD', fontsize=30)
plt.xticks(fontsize=30)
plt.yticks(fontsize=30)
plt.ylabel('Countrate (e-/s)', fontsize=30)
plt.title('Original and Corrected countrates for GD153 in F606W', fontsize=30)
plt.ylim(101000, 104000)
plt.legend(loc=0, fontsize=30)
<matplotlib.legend.Legend at 0x7fe5e8f08750>
6. Redrizzle the corrected FLC frames for each epoch#
The corrected FLC data from each epoch can now be redrizzled to correct for distortion, to remove cosmic rays and bad pixels, and to improve the signal-to-noise in the combined DRC product at each date. (Alternately, the entire set of FLC images may be combined to produce a single DRC image to use for photometry.)
Warning: This cell may take a few minutes to complete.
for asn in glob.glob("*asn.fits"):
astrodrizzle.AstroDrizzle(
asn,
skymethod="match",
skystat="mean",
driz_sep_bits="80",
combine_type="median",
combine_nhigh=1,
driz_cr_snr="3.5 3.0",
driz_cr_scale="2.0 1.5",
final_bits="80",
build=True,
clean=True,
preserve=False,
num_cores=1,
)
Setting up logfile : astrodrizzle.log
AstroDrizzle log file: astrodrizzle.log
AstroDrizzle Version 3.10.0 started at: 20:16:01.689 (02/12/2025)
==== Processing Step Initialization started at 20:16:01.690 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25951624762914 22.031311762662927
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.0852229209196698e-05 1.8306773585025585e-06
CD2_1 CD2_2 : 1.8306773585025585e-06 -1.0852229209196698e-05
NAXIS : 518 546
********************************************************************************
*
* Estimated memory usage: up to 5 Mb.
* Output image size: 518 X 546 pixels.
* Output image file: ~ 3 Mb.
* Cores available: 1
*
********************************************************************************
==== Processing Step Initialization finished at 20:16:01.937 (02/12/2025)
==== Processing Step Static Mask started at 20:16:01.938 (02/12/2025)
==== Processing Step Static Mask finished at 20:16:01.952 (02/12/2025)
==== Processing Step Subtract Sky started at 20:16:01.953 (02/12/2025)
***** skymatch started on 2025-12-02 20:16:01.975416
Version 1.0.11
'skymatch' task will apply computed sky differences to input image file(s).
NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
'MDRIZSKY' header keyword will represent sky value *computed* from data.
----- User specified keywords: -----
Sky Value Keyword: 'MDRIZSKY'
Data Units Keyword: 'BUNIT'
----- Input file list: -----
** Input image: 'ich304prq_flc.fits'
EXT: 'SCI',1; MASK: ich304prq_skymatch_mask_sci1.fits[0]
** Input image: 'ich304q1q_flc.fits'
EXT: 'SCI',1; MASK: ich304q1q_skymatch_mask_sci1.fits[0]
----- Sky statistics parameters: -----
statistics function: 'mean'
lower = None
upper = None
nclip = 5
lsigma = 4.0
usigma = 4.0
binwidth = 0.1
----- Data->Brightness conversion parameters for input files: -----
* Image: ich304prq_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 3.0 [s]
Conversion factor (data->brightness): 212.3487866626491
* Image: ich304q1q_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 3.0 [s]
Conversion factor (data->brightness): 212.3487866626491
----- Computing differences in sky values in overlapping regions: -----
* Image 'ich304prq_flc.fits['SCI',1]' SKY = 3.05637 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.0143932
* Image 'ich304q1q_flc.fits['SCI',1]' SKY = 0 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0
***** skymatch ended on 2025-12-02 20:16:02.105037
TOTAL RUN TIME: 0:00:00.129621
==== Processing Step Subtract Sky finished at 20:16:02.133 (02/12/2025)
==== Processing Step Separate Drizzle started at 20:16:02.134 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25951624762914 22.031311762662927
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.0852229209196698e-05 1.8306773585025585e-06
CD2_1 CD2_2 : 1.8306773585025585e-06 -1.0852229209196698e-05
NAXIS : 518 546
-Generating simple FITS output: ich304prq_single_sci.fits
Writing out image to disk: ich304prq_single_sci.fits
Writing out image to disk: ich304prq_single_wht.fits
-Generating simple FITS output: ich304q1q_single_sci.fits
Writing out image to disk: ich304q1q_single_sci.fits
Writing out image to disk: ich304q1q_single_wht.fits
==== Processing Step Separate Drizzle finished at 20:16:02.474 (02/12/2025)
==== Processing Step Create Median started at 20:16:02.475 (02/12/2025)
reference sky value for image 'ich304prq_flc.fits' is 0.014393150806426957
reference sky value for image 'ich304q1q_flc.fits' is 0.0
Saving output median image to: 'ich3040f0_med.fits'
==== Processing Step Create Median finished at 20:16:02.520 (02/12/2025)
==== Processing Step Blot started at 20:16:02.521 (02/12/2025)
Blot: creating blotted image: ich304prq_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ich304prq_sci1_blt.fits
Writing out image to disk: ich304prq_sci1_blt.fits
Blot: creating blotted image: ich304q1q_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ich304q1q_sci1_blt.fits
Writing out image to disk: ich304q1q_sci1_blt.fits
==== Processing Step Blot finished at 20:16:02.72 (02/12/2025)
==== Processing Step Driz_CR started at 20:16:02.721 (02/12/2025)
Creating output: ich304prq_sci1_crmask.fits
Creating output: ich304q1q_sci1_crmask.fits
==== Processing Step Driz_CR finished at 20:16:02.800 (02/12/2025)
==== Processing Step Final Drizzle started at 20:16:02.801 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25951624762914 22.031311762662927
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.0852229209196698e-05 1.8306773585025585e-06
CD2_1 CD2_2 : 1.8306773585025585e-06 -1.0852229209196698e-05
NAXIS : 518 546
-Generating multi-extension output file: ich3040f0_drc.fits
Deleted all instances of WCS with key A in extensions [1]
Writing out to disk: ich3040f0_drc.fits
==== Processing Step Final Drizzle finished at 20:16:03.432 (02/12/2025)
AstroDrizzle Version 3.10.0 is finished processing at 20:16:03.433 (02/12/2025).
-------------------- --------------------
Step Elapsed time
-------------------- --------------------
Initialization 0.2464 sec.
Static Mask 0.0141 sec.
Subtract Sky 0.1798 sec.
Separate Drizzle 0.3402 sec.
Create Median 0.0455 sec.
Blot 0.1988 sec.
Driz_CR 0.0792 sec.
Final Drizzle 0.6309 sec.
==================== ====================
Total 1.7347 sec.
Trailer file written to: astrodrizzle.log
Setting up logfile : astrodrizzle.log
AstroDrizzle log file: astrodrizzle.log
AstroDrizzle Version 3.10.0 started at: 20:16:03.472 (02/12/2025)
==== Processing Step Initialization started at 20:16:03.474 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.2588967794268 22.031359382197184
CRPIX : 282.5 290.5
CD1_1 CD1_2 : 1.082347140916146e-05 1.9936712504674146e-06
CD2_1 CD2_2 : 1.9936712504674146e-06 -1.082347140916146e-05
NAXIS : 565 581
********************************************************************************
*
* Estimated memory usage: up to 5 Mb.
* Output image size: 565 X 581 pixels.
* Output image file: ~ 3 Mb.
* Cores available: 1
*
********************************************************************************
==== Processing Step Initialization finished at 20:16:03.932 (02/12/2025)
==== Processing Step Static Mask started at 20:16:03.935 (02/12/2025)
==== Processing Step Static Mask finished at 20:16:03.954 (02/12/2025)
==== Processing Step Subtract Sky started at 20:16:03.95 (02/12/2025)
***** skymatch started on 2025-12-02 20:16:03.995920
Version 1.0.11
'skymatch' task will apply computed sky differences to input image file(s).
NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
'MDRIZSKY' header keyword will represent sky value *computed* from data.
----- User specified keywords: -----
Sky Value Keyword: 'MDRIZSKY'
Data Units Keyword: 'BUNIT'
----- Input file list: -----
** Input image: 'ibcda4d0q_flc.fits'
EXT: 'SCI',1; MASK: ibcda4d0q_skymatch_mask_sci1.fits[0]
** Input image: 'ibcda4d6q_flc.fits'
EXT: 'SCI',1; MASK: ibcda4d6q_skymatch_mask_sci1.fits[0]
** Input image: 'ibcda4dcq_flc.fits'
EXT: 'SCI',1; MASK: ibcda4dcq_skymatch_mask_sci1.fits[0]
** Input image: 'ibcda4diq_flc.fits'
EXT: 'SCI',1; MASK: ibcda4diq_skymatch_mask_sci1.fits[0]
----- Sky statistics parameters: -----
statistics function: 'mean'
lower = None
upper = None
nclip = 5
lsigma = 4.0
usigma = 4.0
binwidth = 0.1
----- Data->Brightness conversion parameters for input files: -----
* Image: ibcda4d0q_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 1.0 [s]
Conversion factor (data->brightness): 637.0463599879473
* Image: ibcda4d6q_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 1.0 [s]
Conversion factor (data->brightness): 637.0463599879473
* Image: ibcda4dcq_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 1.0 [s]
Conversion factor (data->brightness): 637.0463599879473
* Image: ibcda4diq_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 1.0 [s]
Conversion factor (data->brightness): 637.0463599879473
----- Computing differences in sky values in overlapping regions: -----
* Image 'ibcda4d0q_flc.fits['SCI',1]' SKY = 68.4183 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.107399
* Image 'ibcda4d6q_flc.fits['SCI',1]' SKY = 10.3393 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.0162301
* Image 'ibcda4dcq_flc.fits['SCI',1]' SKY = 12.4503 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.0195437
* Image 'ibcda4diq_flc.fits['SCI',1]' SKY = 0 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0
***** skymatch ended on 2025-12-02 20:16:04.460451
TOTAL RUN TIME: 0:00:00.464531
==== Processing Step Subtract Sky finished at 20:16:04.510 (02/12/2025)
==== Processing Step Separate Drizzle started at 20:16:04.511 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.2588967794268 22.031359382197184
CRPIX : 282.5 290.5
CD1_1 CD1_2 : 1.082347140916146e-05 1.9936712504674146e-06
CD2_1 CD2_2 : 1.9936712504674146e-06 -1.082347140916146e-05
NAXIS : 565 581
-Generating simple FITS output: ibcda4d0q_single_sci.fits
Writing out image to disk: ibcda4d0q_single_sci.fits
Writing out image to disk: ibcda4d0q_single_wht.fits
-Generating simple FITS output: ibcda4d6q_single_sci.fits
Writing out image to disk: ibcda4d6q_single_sci.fits
Writing out image to disk: ibcda4d6q_single_wht.fits
-Generating simple FITS output: ibcda4dcq_single_sci.fits
Writing out image to disk: ibcda4dcq_single_sci.fits
Writing out image to disk: ibcda4dcq_single_wht.fits
-Generating simple FITS output: ibcda4diq_single_sci.fits
Writing out image to disk: ibcda4diq_single_sci.fits
Writing out image to disk: ibcda4diq_single_wht.fits
==== Processing Step Separate Drizzle finished at 20:16:05.177 (02/12/2025)
==== Processing Step Create Median started at 20:16:05.177 (02/12/2025)
reference sky value for image 'ibcda4d0q_flc.fits' is 0.10739920657268735
reference sky value for image 'ibcda4d6q_flc.fits' is 0.016230068945693298
reference sky value for image 'ibcda4dcq_flc.fits' is 0.01954372089581475
reference sky value for image 'ibcda4diq_flc.fits' is 0.0
Saving output median image to: 'ibcda4010_med.fits'
==== Processing Step Create Median finished at 20:16:05.253 (02/12/2025)
==== Processing Step Blot started at 20:16:05.254 (02/12/2025)
Blot: creating blotted image: ibcda4d0q_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ibcda4d0q_sci1_blt.fits
Writing out image to disk: ibcda4d0q_sci1_blt.fits
Blot: creating blotted image: ibcda4d6q_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ibcda4d6q_sci1_blt.fits
Writing out image to disk: ibcda4d6q_sci1_blt.fits
Blot: creating blotted image: ibcda4dcq_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ibcda4dcq_sci1_blt.fits
Writing out image to disk: ibcda4dcq_sci1_blt.fits
Blot: creating blotted image: ibcda4diq_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: ibcda4diq_sci1_blt.fits
Writing out image to disk: ibcda4diq_sci1_blt.fits
==== Processing Step Blot finished at 20:16:05.746 (02/12/2025)
==== Processing Step Driz_CR started at 20:16:05.747 (02/12/2025)
Creating output: ibcda4d0q_sci1_crmask.fits
Creating output: ibcda4d6q_sci1_crmask.fits
Creating output: ibcda4dcq_sci1_crmask.fits
Creating output: ibcda4diq_sci1_crmask.fits
==== Processing Step Driz_CR finished at 20:16:05.900 (02/12/2025)
==== Processing Step Final Drizzle started at 20:16:05.901 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.2588967794268 22.031359382197184
CRPIX : 282.5 290.5
CD1_1 CD1_2 : 1.082347140916146e-05 1.9936712504674146e-06
CD2_1 CD2_2 : 1.9936712504674146e-06 -1.082347140916146e-05
NAXIS : 565 581
-Generating multi-extension output file: ibcda4010_drc.fits
Deleted all instances of WCS with key A in extensions [1]
Writing out to disk: ibcda4010_drc.fits
==== Processing Step Final Drizzle finished at 20:16:06.705 (02/12/2025)
AstroDrizzle Version 3.10.0 is finished processing at 20:16:06.706 (02/12/2025).
-------------------- --------------------
Step Elapsed time
-------------------- --------------------
Initialization 0.4588 sec.
Static Mask 0.0191 sec.
Subtract Sky 0.5553 sec.
Separate Drizzle 0.6662 sec.
Create Median 0.0758 sec.
Blot 0.4921 sec.
Driz_CR 0.1531 sec.
Final Drizzle 0.8043 sec.
==================== ====================
Total 3.2247 sec.
Trailer file written to: astrodrizzle.log
Setting up logfile : astrodrizzle.log
AstroDrizzle log file: astrodrizzle.log
AstroDrizzle Version 3.10.0 started at: 20:16:06.75 (02/12/2025)
==== Processing Step Initialization started at 20:16:06.752 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25946702548492 22.030300403211356
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.1001745311092143e-05 -2.8958333815648194e-07
CD2_1 CD2_2 : -2.8958333815648194e-07 -1.1001745311092143e-05
NAXIS : 518 546
********************************************************************************
*
* Estimated memory usage: up to 5 Mb.
* Output image size: 518 X 546 pixels.
* Output image file: ~ 3 Mb.
* Cores available: 1
*
********************************************************************************
==== Processing Step Initialization finished at 20:16:06.993 (02/12/2025)
==== Processing Step Static Mask started at 20:16:06.994 (02/12/2025)
==== Processing Step Static Mask finished at 20:16:07.005 (02/12/2025)
==== Processing Step Subtract Sky started at 20:16:07.006 (02/12/2025)
***** skymatch started on 2025-12-02 20:16:07.027245
Version 1.0.11
'skymatch' task will apply computed sky differences to input image file(s).
NOTE: Computed sky values WILL NOT be subtracted from image data ('subtractsky'=False).
'MDRIZSKY' header keyword will represent sky value *computed* from data.
----- User specified keywords: -----
Sky Value Keyword: 'MDRIZSKY'
Data Units Keyword: 'BUNIT'
----- Input file list: -----
** Input image: 'idbha6ntq_flc.fits'
EXT: 'SCI',1; MASK: idbha6ntq_skymatch_mask_sci1.fits[0]
** Input image: 'idbha6nyq_flc.fits'
EXT: 'SCI',1; MASK: idbha6nyq_skymatch_mask_sci1.fits[0]
----- Sky statistics parameters: -----
statistics function: 'mean'
lower = None
upper = None
nclip = 5
lsigma = 4.0
usigma = 4.0
binwidth = 0.1
----- Data->Brightness conversion parameters for input files: -----
* Image: idbha6ntq_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 3.0 [s]
Conversion factor (data->brightness): 212.3487866626491
* Image: idbha6nyq_flc.fits
EXT = 'SCI',1
Data units type: COUNTS
EXPTIME: 3.0 [s]
Conversion factor (data->brightness): 212.3487866626491
----- Computing differences in sky values in overlapping regions: -----
* Image 'idbha6ntq_flc.fits['SCI',1]' SKY = 0 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0
* Image 'idbha6nyq_flc.fits['SCI',1]' SKY = 0.535884 [brightness units]
Updating sky of image extension(s) [data units]:
- EXT = 'SCI',1 delta(MDRIZSKY) = 0.0025236
***** skymatch ended on 2025-12-02 20:16:07.147090
TOTAL RUN TIME: 0:00:00.119845
==== Processing Step Subtract Sky finished at 20:16:07.177 (02/12/2025)
==== Processing Step Separate Drizzle started at 20:16:07.178 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25946702548492 22.030300403211356
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.1001745311092143e-05 -2.8958333815648194e-07
CD2_1 CD2_2 : -2.8958333815648194e-07 -1.1001745311092143e-05
NAXIS : 518 546
-Generating simple FITS output: idbha6ntq_single_sci.fits
Writing out image to disk: idbha6ntq_single_sci.fits
Writing out image to disk: idbha6ntq_single_wht.fits
-Generating simple FITS output: idbha6nyq_single_sci.fits
Writing out image to disk: idbha6nyq_single_sci.fits
Writing out image to disk: idbha6nyq_single_wht.fits
==== Processing Step Separate Drizzle finished at 20:16:07.522 (02/12/2025)
==== Processing Step Create Median started at 20:16:07.52 (02/12/2025)
reference sky value for image 'idbha6ntq_flc.fits' is 0.0
reference sky value for image 'idbha6nyq_flc.fits' is 0.0025236010551451834
Saving output median image to: 'idbha6040_med.fits'
==== Processing Step Create Median finished at 20:16:07.565 (02/12/2025)
==== Processing Step Blot started at 20:16:07.566 (02/12/2025)
Blot: creating blotted image: idbha6ntq_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: idbha6ntq_sci1_blt.fits
Writing out image to disk: idbha6ntq_sci1_blt.fits
Blot: creating blotted image: idbha6nyq_flc.fits[sci,1]
Using default C-based coordinate transformation...
-Generating simple FITS output: idbha6nyq_sci1_blt.fits
Writing out image to disk: idbha6nyq_sci1_blt.fits
==== Processing Step Blot finished at 20:16:07.763 (02/12/2025)
==== Processing Step Driz_CR started at 20:16:07.763 (02/12/2025)
Creating output: idbha6ntq_sci1_crmask.fits
Creating output: idbha6nyq_sci1_crmask.fits
==== Processing Step Driz_CR finished at 20:16:07.833 (02/12/2025)
==== Processing Step Final Drizzle started at 20:16:07.833 (02/12/2025)
WCS Keywords
Number of WCS axes: 2
CTYPE : 'RA---TAN' 'DEC--TAN'
CUNIT : 'deg' 'deg'
CRVAL : 194.25946702548492 22.030300403211356
CRPIX : 259.0 273.0
CD1_1 CD1_2 : 1.1001745311092143e-05 -2.8958333815648194e-07
CD2_1 CD2_2 : -2.8958333815648194e-07 -1.1001745311092143e-05
NAXIS : 518 546
-Generating multi-extension output file: idbha6040_drc.fits
Deleted all instances of WCS with key A in extensions [1]
Writing out to disk: idbha6040_drc.fits
==== Processing Step Final Drizzle finished at 20:16:08.454 (02/12/2025)
AstroDrizzle Version 3.10.0 is finished processing at 20:16:08.455 (02/12/2025).
-------------------- --------------------
Step Elapsed time
-------------------- --------------------
Initialization 0.2408 sec.
Static Mask 0.0111 sec.
Subtract Sky 0.1707 sec.
Separate Drizzle 0.3444 sec.
Create Median 0.0419 sec.
Blot 0.1971 sec.
Driz_CR 0.0691 sec.
Final Drizzle 0.6206 sec.
==================== ====================
Total 1.6958 sec.
Trailer file written to: astrodrizzle.log
drcs = ['ibcda4010_drc.fits', 'ich3040f0_drc.fits', 'idbha6040_drc.fits']
drcents = [(327.9, 341.4), (251.1, 350.0), (266.6, 273.6)]
6.1 Recalculate photometry on the new DRC frames#
We now perform the photometry on the drizzled (DRC) products, using similar techniques as in Section 4.1. Drizzled images are in units of electrons per second are already corrected for distortion, so we no longer need to divide by the exposure time or apply the PAM prior to computing photometry.
phots = []
mjds = []
pfls = []
for i, drc in enumerate(drcs):
data = fits.getdata(drc, ext=1)
mjds.append(np.mean(df.query('Epoch == {}'.format(i+1))['MJD']))
pfls.append(df.query('Epoch == {}'.format(i+1))['PHOTFLAM'].values[0])
positions = drcents[i]
aperture = CircularAperture(positions, ap)
annulus_aperture = CircularAnnulus(positions, r_in=skyrad[0], r_out=skyrad[1])
annulus_masks = annulus_aperture.to_mask(method='center')
annulus_data = annulus_masks.multiply(data)
mask = annulus_masks.data
annulus_data_1d = annulus_data[mask > 0]
mean_sigclip, _, _ = sigma_clipped_stats(annulus_data_1d)
background = mean_sigclip * aperture.area
apers = [aperture, annulus_aperture]
phot_table = aperture_photometry(data, apers)
final_sum = phot_table['aperture_sum_0'] - background
phots.append(final_sum[0])
mags = -21.1 - 2.5*np.log10(pfls[0]) - 2.5*np.log10(phots) - 2.5*np.log10(1./EE_r10)
print(mags)
[13.60497411 13.60317381 13.60501243]
6.2 Plot DRC countrate vs date#
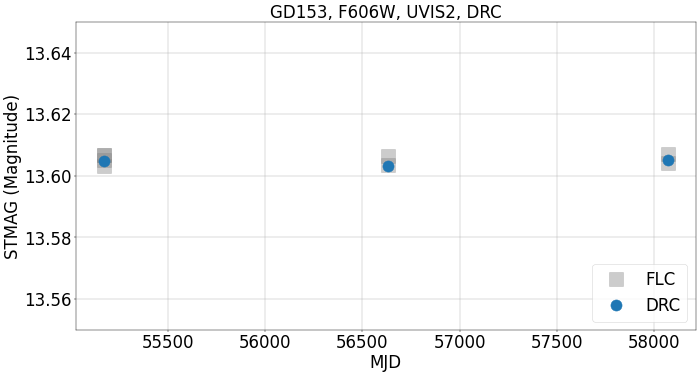
The corrected photometric countrate from the DRC and the recomputed magnitudes are plotted below. Note how they agree with the results from the previous step using the FLC and the PAM.
fig = plt.figure(figsize=(20, 10), dpi=40)
plt.plot(df['MJD'], df['Phot-eq'], 'd', markersize=20, color='grey', label='FLC photometrically equalized', alpha=0.5)
plt.plot(mjds, phots, 'o', markersize=20, label='DRC photometry')
plt.grid()
plt.xlabel('MJD', fontsize=30)
plt.xticks(fontsize=30)
plt.yticks(fontsize=30)
plt.ylabel('Countrate (e-/s)', fontsize=30)
plt.title('GD153, F606W, UVIS2, DRC', fontsize=30)
plt.ylim(101000, 104000)
plt.legend(loc=4, fontsize=30)
<matplotlib.legend.Legend at 0x7fe5e8cf6f90>
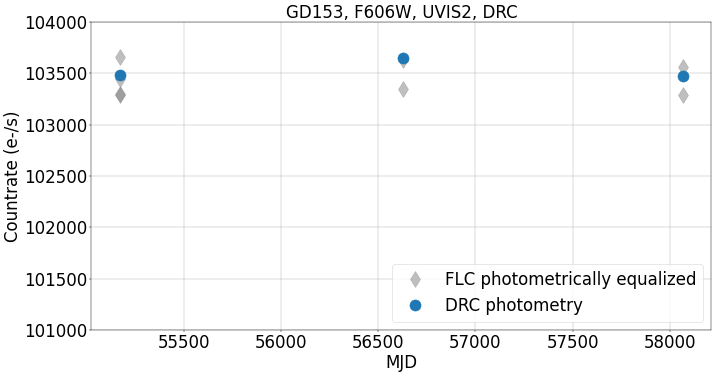
fig = plt.figure(figsize=(20, 10), dpi=40)
plt.plot(df['MJD'], df['STMags'], 's', markersize=25, label='FLC', alpha=0.4, color='Grey')
plt.plot(mjds, mags, 'o', markersize=20, label='DRC')
plt.grid()
plt.ticklabel_format(useOffset=False)
plt.xlabel('MJD', fontsize=30)
plt.xticks(fontsize=30)
plt.yticks(fontsize=30)
plt.ylabel('STMAG (Magnitude)', fontsize=30)
plt.ylim(13.55, 13.65)
plt.title('GD153, F606W, UVIS2, DRC', fontsize=30)
plt.legend(loc=4, fontsize=30)
<matplotlib.legend.Legend at 0x7fe5e8cf5bd0>
7. Conclusions#
Thank you for walking through this notebook. Now using WFC3 data, you should be more familiar with:
Computing aperture photometry and magnitudes on:
FLC frames using new time-dependent photometry keywords.
FLC frames with equalized countrate values.
DRC frames produced from corrected FLCs.
Congratulations, you have completed the notebook!#
Additional Resources#
Below are some additional resources that may be helpful. Please send any questions through the HST Helpdesk.
-
see section 9.5.2 for reference to this notebook
About this Notebook#
Authors: Harish Khandrika, Jennifer Mack; WFC3 Instrument Team
Updated on: 2021-09-10
Citations#
If you use numpy, astropy, drizzlepac, and photutils for published research, please cite the
authors. Follow these links for more information about citing the libraries below:


